ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
- Biomedical Research (2015) Volume 26, Issue 2
Genotype distribution of extended Spectrum ?-Lactamase producing Escherichia coli and Klebsiella pneumoniae.
Department of Microbiology, Faculty of Medicine, Selcuk University, Konya, Turkey
- *Corresponding Author:
- Hatice Turk Dagi
Department of Microbiology
Faculty of Medicine Selcuk University, Konya Turkey
Accepted September 12 2014
Extended-spectrum beta-lactamase (ESBL) production is the most important cause of betalactam resistance in Gram-negative bacteria. Although it may also be found in other Gramnegative bacteria, ESBL is most commonly produced by Escherichia coli and Klebsiella pneumoniae strains. In this study, we aimed to investigate the distribution of β-lactamase genes in ESBL-producing E. coli and K. pneumoniae strains. One hundred and twenty isolates of E. coli and K. pneumoniae isolated from clinical samples were used in this study. The identification and the antibiotic susceptibility tests were performed by VITEK 2 system in accordance with the manufacturer’s instructions. ESBL production was determined accoring to Clinical and Laboratory Standards Institute guidelines. The DNA isolation was performed with a commercial kit following company recommendations. blaTEM, blaSHV and blaCTX-M genes were amplified by multiplex PCR with specific primers. Of the 120 isolates collected, 84 isolates were of E. coli and 36 isolates were of K. pneumoniae. blaTEM gene was the most prevalent type (85.8%) followed by blaCTX-M (83.3%) and blaSHV (24.2%). No blaSHV gene was detected in the E. coli strains. Among 120 ESBL-producing strains, 10.8% were susceptible to cefepime, 10.0% to ceftazidime, while 5.0% to ceftriaxone. In conclusion, blaTEM gene was the most frequently encountered ESBL of E. coli and K. pneumonia in our hospital. Further molecular surveillance and epidemiological studies of such resistant bacteria are recommended for monitoring and controlling the spread of ESBL producing strains.
Keywords
Extended-spectrum beta-lactamase, Escherichia coli, Klebsiella pneumoniae, TEM, SHV, CTX-M
Introduction
Gram-negative bacteria are more resistant to antibiotics than Gram-positive bacteria due to their outer membrane structure of the cell walls, and acquire the multiple resistances with the transfer of genetic materials and/or the selective pressure of the antibiotics. Extended-spectrum beta-lactamase (ESBL) production is the most prominent cause of beta-lactam resistance in Gram-negative bacteria that mediate a resistance to penicillins, cephalosporins and monobactams [1]. Recently, infections caused by ESBL producers have become an emerging public health concern worldwide because ESBL-producing isolates exhibit multidrug resistant phenotype, including the resistance to aminoglycosides and fluoroquinolones, and limit the treatment options available [2].
TEM, SHV and CTX-M, which are the three basic types of ESBLs, have been isolated worldwide. Of over 300 variants of ESBL have been defined and the strains expressing CTX-M-type ESBLs have rapidly spread over the past decade. The genes encoding ESBLs, usually carried on plasmids, facilitate their spread among Gramnegative bacteria [3,4]. Although it may also be determined in other Gram-negative bacteria, ESBLs are most commonly produced by E. coli and K. pneumoniae strains.
Detection of ESBL is initially based on phenotypic tests, such as the double-disc synergy test and combined disc method. However, these tests are time-consuming and inhibited by the AmpC β-lactamases. Over the past years, PCR has replaced traditional phenotypic methods [5]. The molecular detection of the common ESBL genes and the antimicrobial resistance can provide reliable information about their epidemiology. In this study, we aimed to investigate the distribution of blaTEM, blaSHV and blaCTX-M genes in ESBL- producing E. coli and K. pneumoniae strains by multiplex PCR.
Material and Methods
One hundred and twenty isolates of E. coli and K. pneumoniae used for this study were isolated from clinical samples in the Microbiology Laboratory of Selcuk University, Faculty of Medicine, in the year 2013. The identification and the antibiotic susceptibility tests were performed by VITEK 2 system (bioMerieux, France) in accordance with the manufacturer’s instructions. ESBL production was determined by double-disc synergy test according to Clinical and Laboratory Standards Institute (CLSI) guidelines [6]. E. coli ATCC 25922 and K. pneumoniae ATCC 700603 were used as the control strains..
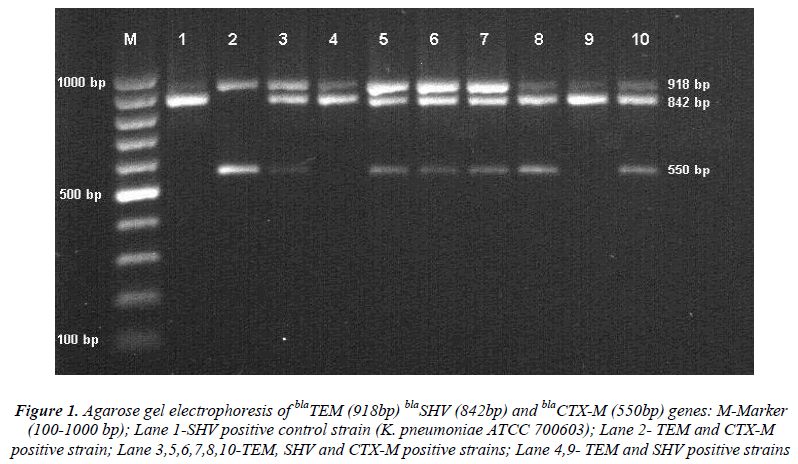
The DNA isolation was performed with a commercial kit (Qiagen, Germany) following the company recommendations. Multiplex PCR was carried out to detect blaTEM,blaSHV and blaCTX-M genes with specific primers. PCR amplification steps included an initial denaturation at 95°C for two minutes and following 30 cycles at 95°C for 45 sec, annealing at 62°C for 45 sec, elongation at 72°C for one minute and a final elongation at 72°C for five minutes. The PCR products were analyzed on 2% agarose gel and visualized with UV light transilluminator staining 0.5μg/mL ethidium bromide. Presence of bands at 918 bp, 842 and 550 bp was considered positive for the blaTEM,S blaSHV and blaCTX-M genes respectively [7].
Results
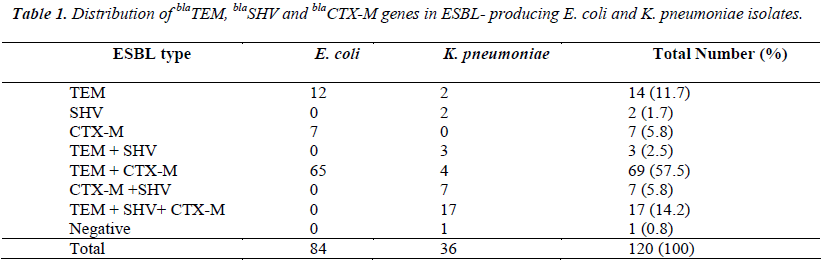
Of the 120 collected isolates, 84 isolates were of E. coli, and 36 isolates were of K. pneumoniae. Single gene was detected in 23 (19.2%) isolates, various combinations of the genes were found in 96 (80%) isolates. blaTEM gene was detected in 103 isolates (85.8%), blaCTX-M in 100 (83.3%), and blaSHV in 29 (24.2%). No blaSHV gene was detected in E. coli strains. All isolates were susceptible to colistin and tigecycline. Among 120 ESBL-producing strains, 98% were susceptible to meropenem, 97% to ertapenem, 76% to amikacin, 52% to piperacillintazobactam, 46% to gentamicin, 33% to trimethoprimsulfamethoxazole, 18% to levofloxacin, 10.8% to cefepime, 10.0% to ceftazidime, and 5.0% to ceftriaxone. However, all strains were resistant to ampicillin and amoxicillin clavulanate.
Figure 1: Agarose gel electrophoresis of blaTEM (918bp) blaSHV (842bp) and blaCTX-M (550bp) genes: M-Marker (100-1000 bp); Lane 1-SHV positive control strain (K. pneumoniae ATCC 700603); Lane 2- TEM and CTX-M positive strain; Lane 3,5,6,7,8,10-TEM, SHV and CTX-M positive strains; Lane 4,9- TEM and SHV positive strains
Discussion
The ESBL-producing bacteria have been dramatically spreading worldwide, requiring continuous monitoring systems and effective infection control measures. The use of antibiotics, particularly third-generation cephalosporins, and the transfer among hospitals are risk factors for acquisition of ESBL-producing bacteria [2,3]. Treat-ment options of infections caused by ESBL-producing isolates are becoming increasingly limited because ESBL-producing bacteria are generally resistant to cephalos-porins and aminoglycosides [8,9]. In aggrement with this, most of the strains are susceptible to meropenem, ertapenem and amikacin, but the resistance rates against third-generation cephalosporins, trimethoprim-sulfamethoxazole, gentamycin and levofloxacin are very high in our study.
The prevalence of ESBL producers and the distribution of ESBL genotypes vary markedly in different geographical areas. Nowadays, the CTX-M type β lactamase is predominant among ESBLs all over the world [10-12]. Similarly, CTX-M-type is the most common β-lactamase found in Turkey too. According to the results of the multicenter ‘HITIT Study’ in Turkey, during 2004 and 2005, the rate of CTX-M-type β-lactamase was 71.4%, TEM 49.4% and SHV 46.7% in E. coli and K. pneumoniae isolates [13]. In another study, blaCTX-M genes were detected in 167 out of 200 (83.5%) Enterobacteriaceae isolates obtained from various clinical samples in Istanbul [14]. In a nationwide study from Turkey, CTX-M1 was the most prevalent (83.18%) gene followed by TEM-1b (44.09%), CTX-M2 (31.81%) and SHV-11 (1.81%) in E. coli isolates [15]. In a different study, the percentage of CTX-M, TEM and SHV in E. coli were determined to be 93%, 64% and 11%, respectively in urinary isolates acquired from community [16]. In contrast to these studies, among non-duplicate 41 ESBL positive isolates of E. coli, Klebsiella spp. and Enterobacter spp., 35 (85.4%) had blaTEM , 33 (80.5%) blaSHV and 20 (48.8%) blaCTXM [17]. In another study, SHV (92.9%) was detected as the most prevalent ESBL type, and TEM and CTX-M (64.3%) were equivalent in K. pneumoniae strains [18]. In our study, TEM (85.8%) and CTX-M (83.3%) types were approximately at the same rates, while SHV was detected in 24.2% of the strains and all of them were K. pneumonia strains. These differences may be caused by several factors such as type and number of the samples, number of the isolates studied, genus and species of the isolates.
blaTEM is a broad spectrum β-lactamase that is always combined with CTX-M in the same plasmid and the combinations of these genes are frequently seen in the ESBL producing strains [10]. Sharma et al. [19] observed that 77.5% of isolates carried more than one type of β lactamase genes, while 32.5% of isolates harbored three β lactamase genes, 22.5% TEM and CTX-M, 15% SHV and CTX-M, and 7.5% TEM and SHV in ESBL producing E. coli and Klebsiella spp. isolates. In our study, while single gene was detected in 23 (19.2%) isolates, various combinations of the genes were found in 96 (80%) isolates. The coexistence of the TEM and SHV was 2.5%, TEM and CTX-M 57.5%, CTX-M and SHV 5.8%, and TEM, SHV and CTX-M 14.2%. Alike in a study from Turkey, TEM and CTX-M were found together in 52%, TEM, SHV and CTX-M were 5%, and CTX-M and SHV were 1.8% [16] On the other hand Bali et al., [20] noticed that most of the ESBL isolates (80.8%) carried one type of β lactamase genes.
In conclusion, blaTEM gene was the most frequently encountered ESBL of E. coli and K. pneumonia in our hospital. There was no blaSHV gene in E. coli strains. The detection and identification of beta lactamases are essential for a reliable epidemiological investigation. The molecular surveillance and epidemiological studies of such resistant bacteria are recommended for monitoring and controlling the spread of ESBL producing strains.
Acknowledgements
The authors declare no conflict of interests. A part of this study was accepted as a poster presentation in the 6th Eurasia Congress of Infectious Diseases, (24-27 September 2014,) in Belgrad, Serbia.
References
- Kanamori H, Navarro RB, Yano H, Sombrero LT, Capeding MR, Lupisan SP, Olveda RM, Arai K, Kunishima H, Hirakata Y, Kaku M. Molecular charac- teristics of extended-spectrum β-lactamases in clinical isolates of Enterobacteriaceae from the Philippines. Acta Trop 2011; 120: 140-145.
- Pitout JD, Laupland KB. Extended-spectrum beta- lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis 2008; 8: 159-166
- Kiratisin P, Apisarnthanarak A, Laesripa C, Saifon P. Molecular characterization and epidemiology of ex- tended-spectrum-beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates causing health care-associated infection in Thailand, where the CTX- M family is endemic. Antimicrob Agents Chemother 2008; 52: 2818-2824.
- Silva-Sanchez J, Ulises Garza-Ramos J, Reyna-Flores F, Sánchez-Perez A, Rojas-Moreno T, Andrade- Almaraz V, Pastrana J, Castro-Romero JI, Vinuesa P, Barrios H, Cervantes C. Extended-spectrum β-lactam- ase-producing Enterobacteriaceae causing nosocomial infections in Mexico. A retrospective and multicenter study. Arch Med Res 2011; 42: 156-162.
- Ahmed OI, El-Hady SA, Ahmed TM, Ahmed IZ. De- tection of bla SHV and bla CTX-M genes in ESBL pro- ducing Klebsiella pneumoniae isolated from Egyptian patients with suspected nosocomial infections. The Egyptian Journal of Medical Human Genetics 2013; 14: 277-283.
- Clinical and Laboratory Standards Institute (CLSI).Performance standards for antimicrobial susceptibility testing. CLSI document M100-S16, Wayne, Pennsyl- vania USA, 2011.
- Kaur M, Aggarwal A. Occurrence of the CTX-M, SHV and the TEM Genes among the Extended Spectrum β- Lactamase Producing Isolates of Enterobacteriaceae in a Tertiary Care Hospital of North India. J Clin Diagn Res 2013; 7: 642-645.
- Feizabadi MM, Delfani S, Raji N, Majnooni A, Alig- holi M, Shahcheraghi F, Parvin M, Yadegarinia D. Dis- tribution of blaTEM, blaSHV, blaCTX-M Genes Among Clinical Isolates of Klebsiella pneumoniae at Lab- bafinejad Hospital, Tehran, Iran. Microb Drug Resist 2010; 16: 49-53.
- Chong Y, Yakushiji H, Ito Y, Kamimura T. Clinical and molecular epidemiology of extended-spectrum β- lactamase-producing Escherichia coli and Klebsiella pneumoniae in a long-term study from Japan. Eur J Clin Microbiol Infect Dis 2011; 30: 83-87.
- Bonnet R. Growing group of extended-spectrum beta- lactamases: the CTX-M enzymes. Antimicrob Agents Chemother 2004; 48: 1-14.
- Livemore DM, Canton R, Gniadkowski MP, Rossolini GM, Arlet G, Ayala J, Coque TM, Kern-Zdanowicz I, Luzzaro F, Poirel L, Woodford N. CTX-M: changing the face of ESBLs in Europe. J Antimicrob Chemother 2007; 59: 165-174.
- Yano H, Uemura M, Endo S, Kanamori H, Inomata S, Kakuta R, Ichimura S, Ogawa M, Shimojima M, Ishi- bashi N, Aoyagi T, Hatta M, Gu Y, Yamada M, To- kuda K, Kunishima H, Kitagawa M, Hirakata Y, Kaku M. Molecular characteristics of extended-spectrum β- lactamases in clinical isolates from Escherichia coli at a Japanese tertiary hospital. PLoS One 2013; 8(5): e64359. doi: 10.1371/journal.pone.0064359.
- Gur D, Gulay Z, Akan OA, Aktaş Z, Kayacan CB, Cakici O, Eraç B, Gültekin M, Ogunç D, Soyletir G, Unal N, Uysal S. Resistance to newer beta-lactams and related ESBL types in gram-negative nosocomial iso- lates in Turkish hospitals: results of the multicentre HITIT study. Mikrobiyol Bul 2008; 42: 537-544.
- Bayraktar B, Toksoy B, Bulut E. Detection of blaCTX-M beta-lactamase genes in extended-spectrum beta- lactamase producing gram-negative bacteria. Mikrobi- yol Bul 2010; 44: 187-196.
- Copur Cicek A, Saral A, Ozad Duzgun A, et al. Na- tionwide study of Escherichia coli producing extended- spectrum β-lactamases TEM, SHV and CTX-M in Tur- key. J Antibiot 2013; 66: 647-650.
- Daglar D, Ongut G, Ogunç D, et al. Investigation of ESBL types in community acquired urinary Es- cherichia coli isolates by isoelectric focusing and po- lymerase chain reaction. Mikrobiyol Bul 2010; 44: 367-374.
- Eser OK, Ergin A, Hascelik G. Occurrence and charac- terisation of CTX-M enzymes in Enterobacteriac- eae isolated from intensive care units of a Turkish Uni- versity hospital. Indian J Med Microbiol 2013; 31: 415- 416.
- Oksuz L, Gurler N. Typing of extended-spectrum beta- lactamases in Escherichia coli and Klebsiella spp. strains and analysis of plasmid profiles. Mikrobiyol Bul 2009; 43: 183-194.
- Sharma M, Pathak S, Srivastava P. Prevalence and an- tibiogram of Extended Spectrum β-Lactamase (ESBL) producing Gram negative bacilli and further molecular characterization of ESBL producing Escherichia coli and Klebsiella spp. J Clin Diagn Res 2013; 7: 2173- 2177.
- Bali EB, Acık L, Sultan N. Phenotypic and molecular characterization of SHV, TEM, CTX-M and extended- spectrum-lactamase produced by Escherichia coli, Aci- nobacter baumannii and Klebsiella isolates in a Turk- ish hospital. African Journal of Microbiology Research 2010; 4: 650-654.