ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2016) Volume 27, Issue 4
Effects of edaravone on hypoxic human astrocytes revealed by a proteomic approach
Taigen Sase1,2, Mitsumi Arito1*, Hidetaka Onodera2, Manae S. Kurokawa1, Yuichiro Tanaka2 and Tomohiro Kato1
1Clinical Proteomics and Molecular Medicine, St. Marianna University Graduate School of Medicine, 2-16-1 Sugao, Miyamae, Kawasaki, Kanagawa 216-8511, Japan
2Department of Neurosurgery, St. Marianna University Graduate School of Medicine, 2-16-1 Sugao, Miyamae, Kawasaki, Kanagawa 216-8511, Japan
- *Corresponding Author:
- Mitsumi Arito
PhD Clinical Proteomics and Molecular Medicine
St. Marianna University Graduate School of Medicine
Japan
Accepted date: March 25, 2016
Introduction: Edaravone (3-methyl-1-phenyl-2-pyrazolin-5-one) of a free radical scavenger has been widely used in treatment of acute ischemic stroke. However, effects of edaravone on hypoxic astrocytes have not been fully understood. Therefore, we investigated the effects by proteomic analysis.
Methods: We cultured human astrocytes under hypoxia and normoxia with or without edaravone. Then, we compared protein profiles of the astrocytes using 2-dimensional differential gel electrophoresis (2DDIGE). Further, we identified proteins affected by hypoxia and/or edaravone by mass spectrometry.
Results: We detected 507 protein spots by 2D-DIGE. Among them, intensity of 22 spots showed more than 1.5-fold increase or less than 1/1.5-fold decrease with statistical significance (p<0.05) by hypoxia. We were able to identify 6 out of the 22 protein spots (increased: alpha-enolase (ENO1, from 2 spots) and ephrin-B2, decreased: adenosine triphosphate synthase subunit alpha (ATPA), mitochondrial heat shock 70 kDa protein 1A/1B, and ferritin light chain). In addition, in 61 out of the 507 spots, the spot intensity showed more than 1.3-fold increase or less than 1/1.3-fold decrease with statistical significance (p<0.05) by hypoxia. In 9 out of the 61 spots, the hypoxia-induced intensity alteration was suppressed by the simultaneous addition of edaravone. Three out of the 9 protein spots were identified (ENO1, ATPA, and rab-interacting lysosomal protein-like protein 1).
Conclusion: Edaravone suppressed a part of molecular responses of human astrocytes to hypoxia. Our data would support use of edaravone in acute ischemic stroke.
Keywords
Astrocyte, Edaravone, Hypoxia, Proteomics
Introduction
The neurovascular unit (NVU) is a functionally integrated structure that is mainly composed of neuronal cells, astrocytes, and microvascular endothelial cells [1,2]. NVU is thought to regulate the interaction between brain circulation and neuronal functions [1,2]. Thereby, sparing of the NVU functions would be important in treatment of acute ischemic stroke [3]. In brain parenchyma, endothelial cells of blood vessels are connected to astrocytes via the foot processes of astrocytes [4]. Similarly, neurons are also connected to astrocytes via other foot processes of astrocytes [1]. Thus, astrocytes play a central role in NVU as a regulator of the interactions between blood vessels and neurons. Furthermore, astrocytes also play a role in maintenance of homeostasis and regulation of responses of the central nervous system to injury [5,6]. From these backgrounds, sparing of astrocyte functions would be of critical importance for sparing of NVU functions in the treatment of acute ischemic stroke.
Edaravone (3-methyl-1-phenyl-2-pyrazolin-5-one) of a free radical scavenger has been widely used in the treatment of acute ischemic stroke [7,8]. Use of edaravone reduces infarction areas and ameliorates neurological deficits in patients with acute ischemic stroke [9]. Similarly, use of edaravone was suggested to improve outcomes of the patients with acute ischemic stroke who were treated with tissue plasminogen activator [10]. Edaravone is thought to work mainly as an antioxidant to inhibit lipid peroxidation [11-13]. However other effects of edaravone have been reported. For example, we reported that edaravone promoted formation of the tight junction between microvascular endothelial cells partly via the down-regulation of monocyte chemoattractant protein-1 secretion [14]. Furthermore, in hypoxic astrocytes, edaravone has been reported to down-regulate hypoxiainducible factor (HIF)-1 and inhibit the production of vascular endothelial growth factor (VEGF) that promotes brain edema [15,16]. However, effects of edaravone on hypoxic astrocytes have not been fully understood. Based on these backgrounds, we here attempted to investigate effects of edaravone on human brain astrocytes in the hypoxic condition by 2- dimensional fluorescence difference gel electrophoresis (2DDIGE) of a comprehensive protein profile analysis.
Materials and Methods
Cell culture
Human astrocytes (Lonza Group Ltd., Basel, Switzerland) were cultured in Astrocyte Basal Medium (ABMTM, Lonza Walkersville, Inc., USA) containing 0.1% recombinant human epidermal growth factor, 0.25% insulin, 3% fetal bovine serum, 0.1% ascorbic acid, 1% L-Glutamine, and 0.1% GA-1000 (AGMTM BulletKitTM, AGMTM plus SingleQuotsTM of Growth Supplements, Lonza Walkersville, Inc., USA). The astrocytes were cultured at 37 degrees centigrade in humidified air with 5% CO2.
Treatment of astrocytes with edaravone
Astrocytes were cultured under hypoxia and normoxia condition with or without 10 μM edaravone (Mitsubishi Tanabe Pharma Corporation, Osaka, Japan). The hypoxia condition (1% O2, 5% CO2, and 94% N2) was achieved using a hypoxic chamber (Hypoxia Chamber, Stemcell Technologies).
RNA extraction and reverse transcription-PCR (RTPCR)
RNA extraction and RT-PCR were performed as described previously [17,18]. Extraction and purification of RNA from the astrocytes and reverse transcription of the RNA samples were performed using RNeasy (Qiagen, Venlo, The Netherlands) and High Capacity cDNA Reverse Transcription Kits (Life Technologies, Rockville, MD, USA), respectively. Then, 2 μg of total RNA-derived cDNA was mixed with 1 μM each of the forward and reverse primers and Premix TaqTM (Ex TaqTM Version 2.0, Takara, Shiga, Japan) and subjected to PCR. Nucleotide sequences of the primers for the amplification of a VEGF fragment and β-actin (ACTB) fragment are as follows: for VEGF; 5’- GCCTCCGAAACCATGAACTTTCTGCTG and 5’- TGGTGATGTTGGACTCCTCA, and for ACTB; 5’- AGGCACCAGGGCGTGAT and 5’- TGCTCCCAGTTGGTGACGAT. The thermal cycle conditions were as follows: for VEGF; 95 for 5 min, 25 cycles (95°C for 15 s, 60°C for 30 s, and 72°C for 20 s), and 72°C for 5 min, and for ACTB; 98°C for 5 min, 25 cycles (98°C for 15 s, 58°C for 30 s, and 72°C for 20 s), and 72°C for 5 min.
Quantitative PCR (qPCR)
qPCR was performed as described previously [18]. qPCR was performed using ABI Prism 7000 Sequence Detection System (Applied BiosystemsTM, Foster city, CA, USA), according manufacturer’s instructions. 1 μg of total RNA-derived cDNA was mixed with TaqMan® Gene Expression Assays (VEGF: Hs00900055_m1, and ACTB: Mm00607939_s1, Applied BiosystemsTM) and TaqMan® Gene Expression Master Mix (Applied BiosystemsTM) and then was subjected to qPCR. The thermal cycle conditions were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s.
2-dimensional fluorescence difference gel electrophoresis analysis (2D-DIGE)
Proteins extracted from the astrocytes were separated by 2DDIGE. 2D-DIGE was performed as described previously [14,19,20]. An internal control sample was prepared by mixing an equal amount of samples from the 4 conditions of normoxia without edaravone, normoxia with edaravone, hypoxia without edaravone, and hypoxia with edaravone. The internal control sample was labeled with Cy3 (Cy Dye DIGE Saturation dye; GE Healthcare, Buckinghamshire, UK). Similarly, each of the treated protein samples was labeled with Cy5. Then, 2.5 μg of the Cy3-labeled internal control sample and 2.5 μg of the Cy5- labeled samples were mixed and applied onto an isoelectric focusing (IEF) gel (pH3-11, GE Healthcare). After IEF, the proteins were further separated by 12.5% SDS-PAGE, separated protein spots were detected using an image analyzer (Typhoon 9400 Imager, GE Healthcare). To compare protein spot intensity among the 4 groups, Cy5-fluorescent intensity of protein spots in each gel was normalized by Cy3-fluorescent intensity of the identical spots by using the Progeneosis software (Nonlinear Dynamics, Newcastle, UK), and the normalized Cy5-intensity was used for the comparison.
Protein identification
Protein identification was performed as described previously [14,19,20]. For identification of proteins, gel fragments that corresponded to protein spots of interest were recovered. Peptides, generated by in-gel digestion with trypsin, were extracted from the gel fragments and were subjected to MALDI-TOF/TOF mass spectrometry (Ultraflex, Bruker Daltonics, Bremen, Germany). The determined peptide masses were compiled to allow searches of the NCBI (National Center for Biotechnology Information) protein database using the Mascot software program (Matrix Svience, London, UK).
Statistical analysis
Statistical significance was calculated by Student’s t-test.
Results
Changes of astrocyte protein profiles by hypoxia
To understand effects of edaravone on hypoxic human astrocytes, we here investigated protein profile changes of human astrocytes by hypoxia and effects of edaravone on the changes using 2D-DIGE.
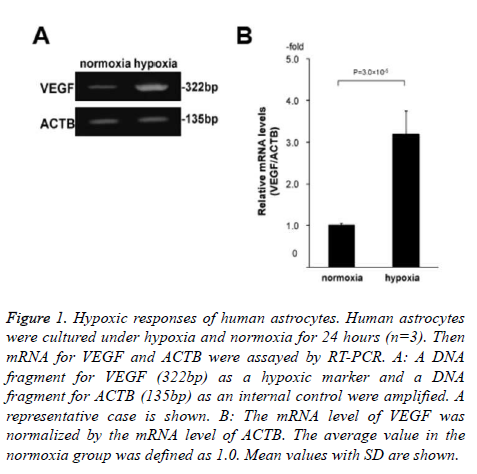
Specifically, we cultured human astrocytes under hypoxia and normoxia for 24 hours with or without 10 μM edaravone (n=3 for each of the 4 conditions; normoxia without edaravone, hypoxia without edaravone, normoxia with edaravone, and hypoxia with edaravone). Before the 2D-DIGE analysis, we confirmed that the astrocytes were exposed to hypoxia by the assessment of mRNA levels of VEGF between the normoxia and hypoxia samples without edaravone. As a result, the levels of VEGF were significantly high in the hypoxia samples compared to the normoxia samples (3.2-folds, Figure 1). This indicated that the astrocytes were exposed to hypoxia condition in our experiment.
Figure 1. Hypoxic responses of human astrocytes. Human astrocytes were cultured under hypoxia and normoxia for 24 hours (n=3). Then mRNA for VEGF and ACTB were assayed by RT-PCR. A: A DNA fragment for VEGF (322bp) as a hypoxic marker and a DNA fragment for ACTB (135bp) as an internal control were amplified. A representative case is shown. B: The mRNA level of VEGF was normalized by the mRNA level of ACTB. The average value in the normoxia group was defined as 1.0. Mean values with SD are shown.
Next, we separated proteins of each sample by 2D-DIGE as shown in figure 2, by which we detected 507 protein spots in total (Table 1). In the comparison of astrocyte protein profiles between normoxia and hypoxia conditions, we found that intensity of 116 out of the 507 protein spots were different with statistical significance as shown in Table 1. Out of the 116 protein spots, 39 protein spots showed more than 1.3-fold higher intensity in the hypoxia condition than in the normoxia condition. 15 out of the 39 protein spots showed more than 1.5- fold higher intensity. Similarly, 22 protein spots showed less than 1/1.3-fold lower intensity in the hypoxia condition than in normoxia condition.
Figure 2. 2D-DIGE detection of proteins affected by edaravone in hypoxic human astrocytes. Human astrocytes were cultured under hypoxia and normoxia with or without 10 μM edaravone for 24 hours (n=3, each). Proteins extracted from each of the astrocyte samples mixed with the internal standard sample were separated by 2DDIGE. MW; molecular weight, pI: isoelectric point.
| Detected spots on 2-DE gels |
hypoxia/normoxia (p<0.05) |
Effects of edaravone (under hypoxia) (p<0.05) |
|||
|---|---|---|---|---|---|
| Total | Difference | Spot count | Difference | Spot count | |
| 507 | 116 | ↑ (1.5 £) | 15 | ↓ | 2 |
| → | 13 | ||||
| ↑ | 0 | ||||
| ↑ (1.3 £) | 39 | ↓ → ↑ |
5 34 0 |
||
| ↓ (£ -1.3) | 22 | ↓ → ↑ |
0 18 4 |
||
| ↓ (£ -1.5) | 7 | ↓ | 0 | ||
| → | 4 | ||||
| ↑ | 3 | ||||
Table 1. Numbers of protein spots whose intensity was altered more than 1.3-fold by hypoxia and the alternation was suppressed by edaravone.
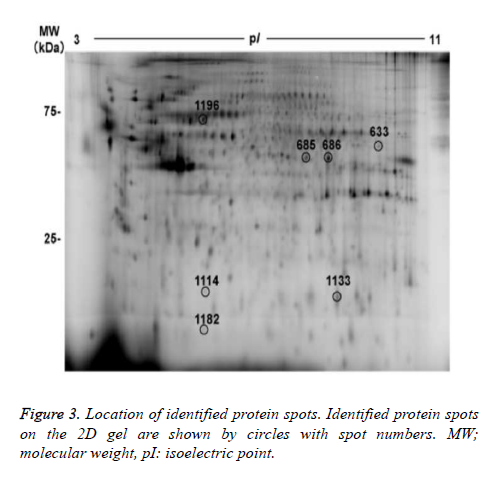
7 out of the 22 protein spots showed less than 1/1.5-fold lower intensity. Focusing on the 22 protein spots that showed more than 1.5- or less than 1/1.5- fold changes, we tried to identify protein names using MALDI-TOF/TOF mass spectrometry. As a result, protein names were identified in 6 protein spots (nos. 633, 685, 686, 1114, 1133, and 1196), as summarized in figure 3 and table 2. Three out of the 6 identified spots (no.685 of alpha-enolase (ENO1), no.686 of ENO1, and no.1113 of ephrin-B2 (EFNB2), showed higher intensity in the hypoxia condition than in the normoxia condition. The remaining 3 spots (no. 633 of adenosine triphosphate synthase subunit alpha (ATPA), no. 1196 of mitochondrial heat shock 70 kDa protein 1A/1B (HSPA1A), and no.1114 of ferritin light chain (FTL)) showed lower intensity in the hypoxia condition than in the normoxia condition.
| Spot no. |
Fold changes (hypoxia/normoxia) |
MW/pI (observed) |
Protein | Accession no. |
MW/pI (calculated) |
Matched peptides |
Coverage (%) |
Sequence confirmed by LID (Mascot ion score) |
|---|---|---|---|---|---|---|---|---|
| 685 | 1.6 | 48.9/7.4 | Alpha-enolase (ENO1_HUMAN) |
GI:4503571 | 47.1/7.01 | 238 | 14 | 33AAVPSGASTGIYEALELR50 (106) 163LAMQEFMILPVGAANFR179 + 2 Oxidation (M) (41) |
| 686 | 1.6 | 48.7/7.8 | Alpha-enolase (ENO1_HUMAN) |
GI:4503571 | 47.1/7.01 | 184 | 22 | 33AAVPSGASTGIYEALELR50 (78) 240VVIGMDVAASEFFR253 + Oxidation (M) (58) |
| 1133 | 1.6 | 16.7/7.9 | Ephrin-B2 (EFNB2_HUMAN) |
GI:4758250 | 36.9/9.04 | 59 | 15 | 182RPELEAGTNGR192 (8) 295TADSVFCPHYEK306 (17) |
| 633 | -1.7 | 53.1/8.7 | ATP synthase subunit alpha, mitochondrial (ATPA_HUMAN) | GI:4757810 | 59.7/9.16 | 106 | 15 | 335EAYPGDVFYLHSR347 (31) 403GIRPAINVGLSVSR416 (17) |
| 1196 | -2.1 | 66.7/5.6 | Heat shock 70 kDa protein1A/1B (HSPA1A_HUMAN) | GI:194248072 | 70.0/5.48 | 158 | 19 | 160DAGVIAGLNVLR171 (21) 172IINEPTAAAIAYGLDR187 (33) 329AQIHDLVLVGGSTR342 (46) |
| 1114 | -3.3 | 18.0/5.7 | Ferritin light chain (FTL_HUMAN) |
GI:20149498 | 20.0/5.52 | 62 | 23 | 155LGGPEAGLGEYLFER169 (19) |
| MW: molecular weight (kDa), pI: isoelectric point, LID: laser-induced dissociation | ||||||||
Table 2. Identification of protein spots whose intensity was altered more than 1.5-fold by hypoxia with statistical significance.
Effects of edaravone on the hypoxia-induced protein profile changes in astrocytes
We then investigated effects of edaravone on the hypoxiainduced protein profile changes in astrocytes. Specifically, we compared the protein profiles between the edaravone-treated and non-treated samples (under hypoxia) using the 2D-DIGE results. Among the 39 protein spots that showed more than 1.3- fold higher intensity by the hypoxia induction, 5 spots showed significant suppression of the intensity increase by the treatment with edaravone (p<0.05, Table 1). Similarly, 22 protein spots that showed less than 1/1.3-fold lower intensity by the hypoxia induction, 4 protein spots showed significant suppression of the intensity decrease by the treatment with edaravone (p<0.05, Table 1). In addition, edaravone did not enhance significantly the hypoxia-induced intensity changes in any of the 116 protein spots.
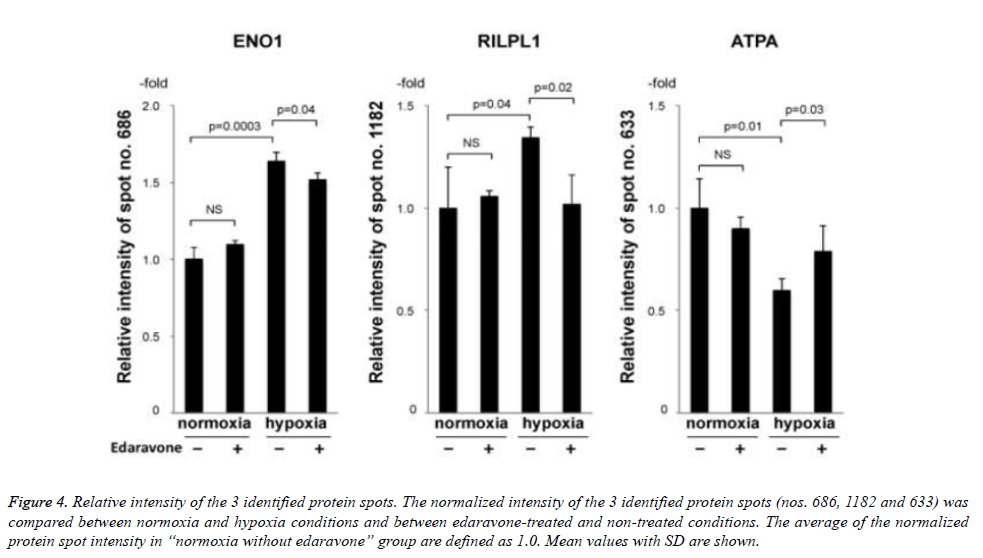
Then, we tried to identify the 9 protein spots in which hypoxiainduced intensity changes were suppressed by the treatment with edaravone, using MALDI-TOF-TOF/MS. As a result, we identified 3 protein spots (no. 686 of ENO1, no. 1182 of rabinteracting lysosomal protein-like protein 1 (RILPL1), and no. 633 of ATPA, Fig. 3 and Table 3). Of note, the intensity of these 3 protein spots was not significantly changed by the treatment with edaravone under normoxia (Figure 4). These data showed that edaravone suppressed the hypoxia-induced change of these protein spots.
Figure 4. Relative intensity of the 3 identified protein spots. The normalized intensity of the 3 identified protein spots (nos. 686, 1182 and 633) was compared between normoxia and hypoxia conditions and between edaravone-treated and non-treated conditions. The average of the normalized protein spot intensity in “normoxia without edaravone” group are defined as 1.0. Mean values with SD are shown.
| Spot no. |
Effects of hypoxia |
Effects of edaravone under hypoxia |
MW/pI (observed) |
Protein | Accession no. |
MW/pI (calculated) |
Matched peptides | Coverage (%) |
Sequence confirmed by LID (Mascot ion score) |
|---|---|---|---|---|---|---|---|---|---|
| a | ↑ | ↓ | 48.7/7.8 | Alpha-enolase (ENO1_HUMAN) |
GI:4503571 | 47.1/7.01 | 184 | 22 | 33AAVPSGASTGIYEALELR50 (78) 240VVIGMDVAASEFFR253 + Oxidation (M) (58) |
| 1182 | ↑ | ↓ | 13.4/5.6 | RILP-like protein 1 (RILPL1_HUMAN) |
GI:195947382 | 47.1/5.13 | 72 | 16 | 201LMKINHDLR209 + Oxidation (M) (24) |
| 633 | ↓ | ↑ | 53.1/8.7 | ATP synthase subunit alpha, mitochondrial (ATPA_HUMAN) | GI:4757810 | 59.7/9.16 | 106 | 15 | 335EAYPGDVFYLHSR347 (31) 403GIRPAINVGLSVSR416 (17) |
| MW; molecular weight (kDa), pI; isoelectric point, LID; laser-induced dissociation | |||||||||
Table 3.Identification of proteins whose intensity changes by hypoxia were suppressed by edaravone with statistical significance.
Discussion
We here found that edaravone suppressed a part of hypoxiainduced protein profile changes of astrocytes and that edaravone did not enhance the hypoxia-induced alteration of the profile. Our data indicate that use of edaravone in the treatment of acute ischemic stroke would be beneficial to protect astrocytes from hypoxia-induced damages. First, intensity of the protein spots of nos. 685, 1133, and 1182, which were assigned to ENO1, EFNB2, and RILPL1 respectively, was increased by hypoxia. The increase of these 3 proteins was in accordance with previous reports. Specifically, expression of ENO1 and EFNB2 was reported to be increased in hypoxic human astrocytes [21] and in hypoxic human trophoblast-derived cell lines [22], respectively. Since EFNB2 is an essential factor for differentiation of vascular endothelial cells into arterial endothelial cells [23], it is reasonable that the expression of EFNB2 was increased by hypoxia to maintain arterial endothelial structures.
Protein spots of nos. 633, 1114, and 1196 whose intensity was decreased by hypoxia were identified as ATPA, FTL, and HSPA1A respectively. ATPA was reported to be decreased by hypoxia in human macrophages [24]. It was reported that the protein level of FTL was increased in lens epithelial cells of mixed breeding dog [25]. The change of FTL expression by hypoxia is likely to be different among tissue type. Then, we thus need to investigate this point in the future. Next, we focused on the effects of edaravone on hypoxic human astrocytes. The intensity of protein spot no. 686, identified as ENO1, was increased by hypoxia and the change was suppressed by the simultaneous treatment with edaravone. ENO1, one of the glycolytic enzymes, converts 2- phosphoglyceric acid to phosphoenolpyruvic acid. The expression of ENO1 was reported to be increased by hypoxia via enhancement of anaerobic metabolism [26,27]. ENO1 was reported to be produced by hypoxia in a HIF-1-dependent manner [21]. Furthermore, it was reported that edaravone inhibited HIF-1 activity [15]. Taking these reports together with ours, edaravone would suppress the expression of ENO1 by the inhibition of HIF-1 activity.
Intensity of protein spot no. 633, identified as ATPA, was decreased by hypoxia and the change was suppressed by the simultaneous treatment with edaravone. ATPA is one of the 5 subunits of ATP synthesize. The ATP synthase contains subunits and among which decrease of ATP synthase subunit (ATPB) in hypoxic rats, decrease of ATPB and ATP synthase subunit in hypoxic mice, and decrease of ATPA and ATPB in hypoxic human macrophages were reported previously [28,29]. We found that edaravone suppressed the decrease of ATPA in hypoxic human astrocytes. It was recently reported that ATPA was decreased by hypoxia at least in part in a HIF-1- independent manner [24]. HIF-1 activity was suppressed by edaravone as above mentioned [15]. Taking these data together with ours, edaravone would suppress expression of ATPA in the hypoxia condition via both HIF-1-dependent and - independent pathways.
It was reported that hypoxia and/or oxidative stress suppressed the synthesis level of ATP in mitochondria, promoting the production of reactive oxygen species (ROS) [30-35]. Therefore, edaravone may suppress ROS production by maintaining the expression of ATPA in the hypoxic condition. The intensity of protein spot no. 1182, identified as RILPL1, was increased by hypoxia and the increase was fully suppressed by the treatment with edaravone. Thereby, RILPL1 would be deeply involved in the functions of edaravone in hypoxia, however, functions of RILPL1 have been little understood. The relationship among hypoxia, RILPL1, and edaravone should be investigated in the future. In conclusion, edaravone suppressed a part of molecular responses of human astrocytes to hypoxia, but did not enhance any of the responses. Although molecular functions of edaravone should be investigated in more detailed, our data here would support the current clinical use of edaravone for the treatment of acute ischemic stroke.
Acknowledgements
We thank Ms. Y. Sawada, Ms. M. Yokoyama and Ms. A. Nozawa for their technical assistance.
References
- del Zoppo GJ. The neurovascular unit in the setting of stroke. J Intern Med 2010; 267: 156-171.
- Lo EH, Dalkara T, Moskowitz MA. Mechanisms, challenges and opportunities in stroke. Nat Rev Neurosci 2003; 4: 399-415.
- Lo EH, Rosenberg GA. The neurovascular unit in health and disease: introduction. Stroke 2009; 40: S2-3.
- Mathiisen TM, Lehre KP, Danbolt NC, Ottersen OP. The perivascular astroglial sheath provides a complete covering of the brain microvessels: an electron microscopic 3D reconstruction. Glia 2010; 58: 1094-1103.
- Liberto CM, Albrecht PJ, Herx LM, Yong VW, Levison SW. Pro-regenerative properties of cytokine-activated astrocytes. J Neurochem 2004; 89: 1092-1100.
- Tsacopoulos M, Magistretti PJ. Metabolic coupling between glia and neurons. J Neurosci 1996; 16: 877-885.
- Abe K, Yuki S, Kogure K. Strong attention of ischemic and postischemic brain edema in rats by a novel free radical scavenger. Stroke 1988; 19: 480-485.
- Albers GW, Goldstein LB, Hess DC, Wechsler LR, Furie KL, Gorelick PB, Hurn P, Liebeskind DS, Nogueira RG, Saver JV, and for the STAIR VII Consortium. Stroke Treatment Academic Industry Roundtable (STAIR) recommendations for maximizing the use of intravenous thrombolytics and expanding treatment options with intra-arterial and neuroprotective therapies. Stroke 2011; 42: 2645-2650.
- Nakase T, Yoshida S, Suzuki, A. Free radical scavenger, edaravone, reduces the lesion size of lacunar infarction in human brain ischemic stroke. BMC Neurol 2011; 11: 39.
- Wada T, Yasunaga H, Inokuchi R, Horiguchi H, Fushimi K, Matsubara T, Nakajima S, Yahagi N. Effects of edaravone on early outcomes in acute ischemic stroke patients treated with recombinant tissue plasminogen activator. J Neurol Sci 2014; 345: 106-111.
- Lukic-Panin V, Deguchi K, Yamashita T, Shang J, Zhang X, Tian F, Liu N, Kawai H, Matsuura T, Abe K. Free radical scavenger edaravone administration protects against tissue plasminogen activator induced oxidative stress and blood brain barrier damage. Curr Neurovasc Res 2010; 7: 319-29.
- Mizuno A, Umemura K, Nakashima M. Inhibitory effect of MCI-186, a free radical scavenger, on cerebral ischemia following rat middle cerebral artery occlusion. Gen Pharmacol 1998; 30: 575-578.
- Watanabe T, Yuki S, Egawa M, Nishi H. Protective effects of MCI-186 on cerebral ischemia: possible involvement of free radical scavenging and antioxidant actions. J Pharmacol Exp Ther 1994; 268: 1597-1604.
- Onodera H, Arito M, Sato T, Ito H, Hashimoto T, Tanaka Y, Kurokawa MS, Okamoto K, Suematsu N, Kato T. Novel effects of edaravone on human brain microvascular endothelial cells revealed by a proteomic approach. Brain Res 2013; 1534: 87-94.
- Ishikawa A, Yoshida H, Metoki N, Toki T, Imaizumi T, Matsumiya T, Yamashita K, Taima K, Satoh K. Edaravone inhibits the expression of vascular endothelial growth factor in human astrocytes exposed to hypoxia. Neurosci Res 2007; 59: 406-412.
- Jośko J, Knefel K. The role of vascular endothelial growth factor in cerebral oedema formation. Folia Neuropathol 2003; 41: 161-166.
- Asano K, Arito M, Kurokawa MS, Omoteyama K, Okamoto K, Suematsu N, Yudoh K, Nakamura H, Beppu M, Kato T. Secretion of inflammatory factors from chondrocytes by layilin signaling. Biochem Biophys Res Commun 2014; 452: 85-90.
- Koizumi H, Arito M, Endo W, Kurokawa MS, Okamoto K, Omoteyama K, Suematsu N, Beppu M, Kato T. Effects of tofacitinib on nucleic acid metabolism in human articular chondrocytes. Mod Rheumatol 2015; 25: 522-7.
- Fukasawa M, Okamoto K, Nakamura M, Mikami K, Shimada S, Tanaka Y, Nagai K, Arito M, Kurokawa MS, Masuko K, Suematsu N, Koizuka I, Kato T. Proteomic analysis of the rat cerebellar flocculus during vestibular compensation. J Vestib Res 2009; 19: 83-94.
- Endo W, Arito M, Sato T, Kurokawa MS, Omoteyama K, Iizuka N, Okamoto K, Suematsu N, Nakamura H, Beppu M, Kato T. Effects of sulfasalazine and tofacitinib on the protein profile of articular chondrocytes. Mod Rheumatol 2014; 24: 844-50.
- Mense SM, Sengupta A, Zhou M, Lan C, Bentsman G, Volsky DJ, Zhang L. Gene expression profiling reveals the profound upregulation of hypoxia-responsive genes in primary human astrocytes. Physiol Genomics 2006; 25: 435-49.
- Chennakesava CS, Di Sabto S, Ziemiecki A, Schneiber H, Andres AC. Differential expression of the receptor tyrosine kinase ephB4 and its ligand ephrin-B2 during human placental development. Placenta 2006; 27: 959-967.
- Adams RH, Wilkinson GA, Weiss C, Diella F, Gale NW, Deutsch U, Risau W, Klein R. Roles of ephrinB ligands and EphB receptors in cardiovascular development: demarcation of arterial/venous domains, vascular morphogenesis, and sprouting angiogenesis. Genes Dev 1999; 13: 295-306.
- Fuhrmann DC, Wittig I, Heide H, Dehne N, Brüne B. Chronic hypoxia alters mitochondrial composition in human macrophages. Biochim Biophys Acta 2013; 1834: 2750-2760.
- Goralska M, Fleisher LN, McGahan MC. Hypoxia induced changes in expression of proteins involved in iron uptake and storage in cultured lens epithelial cells. Exp Eye Res 2014; 125: 135-141.
- Sedoris KC, Thomas SD, Miller DM. Hypoxia induces differential translation of enolase/MBP-1. BMC Cancer 2010; 10: 157.
- Pancholi V. Multifunctional alpha-enolase: its role in diseases. Cell Mol Life Sci 2001; 58: 902-20.
- Li J, Qi Y, Liu H, Cui Y, Zhang L, Gong H, Li Y, Li L, Zhang Y. Acute high-altitude hypoxic brain injury: Identification of ten differential proteins. Neural Regen Res 2013; 8: 2932-2941.
- Dukhande VV, Sharma GC, Lai JCK, Farahani R. Chronic hypoxia-induced alterations of key enzymes of glucose oxidative metabolism in developing mouse liver are mTOR dependent. Mol Cell Biochem 2011; 357: 189-197.
- Hoffman DL, Salter JD, Brookes PS. Response of mitochondrial reactive oxygen species generation to steady-state oxygen tension: implications for hypoxic cell signaling. Am J Physiol Heart Circ Physiol 2007; 292: H101-108.
- Murphy MP. How mitochondria produce reactive oxygen species. Biochem J 2009; 417: 1-13.
- Yang J, Chi Y, Burkhardt BR, Guan Y, Wolf BA. Leucine metabolism in regulation of insulin secretion from pancreatic beta cells. Nutr Rev 2010; 68: 270-279.
- Diaz F, Enríquez JA, Moraes CT. Cells lacking Rieske iron-sulfur protein have a reactive oxygen species-associated decrease in respiratory complexes I and IV. Mol Cell Biol 2012; 32: 415-429.
- Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell 2012; 48: 158-167.
- Huang WY, Jou MJ, Peng TI. mtDNA T8993G mutation-induced F1F0-ATP synthase defect augments mitochondrial dysfunction associated with hypoxia/reoxygenation: the protective role of melatonin. PLoS One 2013; 8: e81546.