ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 15
Effect of targeted mammalian sterile 20-like kinase 1 regulation on proliferation and apoptosis of SW480 colorectal cancer cells
Zhuoqi Liu1#, Hongfei Lu1#, Xiaohong Yang1, Weifeng Zhu1, Huifang Wan2, Lehan Yu2, Baohua Xu3, Xiali Zhang3 and Fusheng Wan1*
1Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Nanchang University, Nanchang, Jiangxi, PR China
2Teaching Center of Medical Experiments, Nanchang University, Nanchang, Jiangxi, PR China
3Scientific Center of Laboratory Animals, Nanchang University, Nanchang, Jiangxi, PR China
#These authors contributed equally to this work
- *Corresponding Author:
- Fusheng Wan
Teaching and Researching Institute of Biochemistry and Molecular Biology
School of Basic Medicine
Nanchang University, PR China
Accepted on June 8, 2017
This study aims to investigate the effects of targeted regulation of Mst1 (Mammalian sterile 20-like kinase 1) expression on the proliferation and apoptosis of SW480 colorectal cancer cells and to elucidate the mechanisms underlying these effects. PolyJetTM in vitro DNA transfection reagent was used to transfect pEGFP-N1-Mst1 into SW480 colorectal cancer cells, and LipofectamineTM2000 was used to transfect Mst1-specific siRNA for the targeted silencing of Mst1. MTT assay was then performed to detect the survival rate of the cells, flow cytometry was used to determine apoptosis, and RT-qPCR and western blot were used to measure the mRNA and protein levels of Mst1, PUMA, p73, p53, YAP (Yes Associated Protein 1), and caspase-3. Compared with the control group, the p-EGFP-Mst1 and p-EGFPMst1+ 5-FU groups showed significantly higher rate of cell proliferation inhibition and apoptosis (P<0.05). The protein levels of MST1, Phospho-YAP1 (Ser127), P73, P53, PUMA, and Caspase-3 were significantly increased (P<0.05), while the protein levels of CTGF (Connective Tissue Growth Factor) and AREG (Amphiregulin) were significantly reduced (P<0.05). In the Mst1-siRNA group, the apoptosis rate was significantly decreased (P<0.01), cell proliferation was accelerated, p73, p53, and PUMA were downregulated, and CTGF, AREG, and YAP were upregulated. The targeted regulation of Mst1 expression significantly affected the proliferation and apoptosis of the SW480 cells. Thus, Mst1 has potential for use as a new target for the prevention and treatment of colorectal cancer.
Keywords
Mammalian sterile 20-like kinase 1, Colorectal cancer, SW480, Cell proliferation.
Introduction
Mammalian sterile 20-like kinase 1/2 (Mst1/2) is a key member of the Hippo signaling pathway in mammals, and plays a role in the regulation of organ size, cell proliferation, apoptosis, and tumor growth inhibition [1-3]. Ste20 (Sterile 20 protein kinase) was first identified from yeast by Leberer et al. [4] and was found to be a key molecule involved in cell proliferation [5] and apoptosis [6,7]. Mst1 is a tumor suppressor gene that can code for Ser/Thr kinase [8]. The Mst1 protein mainly consists of a catalytic domain (N-terminus, 330 amino acids) and a non-catalytic tail (C-terminus), in which the non-catalytic region contains one 63-amino-acid autogenetic inhibitory region, one 56-amino-acid dimerization region (also known as the helical coiling region), and two functional nuclear export signaling peptides [9], the deletion or mutation in which would lead to excessive cell growth and transformation [10]. The Hippo signaling pathway was first identified in Drosophila by Huang et al. [11]. Later, Dong et al. [12] established a Yes Associated Protein 1 (YAP) conditioning transgenic mouse model and determined the Mst1-YAP signaling transduction pathway in mammals. Abnormalities in the Mst1 gene are common in colorectal cancer [13] and liver cancer [14]. Further, Zhou et al. [8] found that mice lacking Mst1/2 could not suppress the proliferation-promoting and anti-apoptotic activities of the oncogene Yap1, thus resulting in the formation of hepatocellular carcinoma. However, when Mst1 was transfected into hepatoma cells, YAP1 loses its proliferation-promoting activity, while its apoptosis-inducing activity is increased. Thus, the tumorigenicity of the hepatoma cells is eliminated, indicating that the expression of Mst1 plays a significant role in tumorigenicity. Previously, we found that the introduction of an exogenous Mst1 gene increased the protein levels of Mst1, Caspase-3, and Phospho-YAP (Ser127) in HepG2 cells, and reduced the protein levels of Connective Tissue Growth Factor (CTGF), Amphiregulin (AREG), and Survivin, thus inhibiting the proliferation of HepG2 cells and promoting apoptosis [15]. Recent studies [16-18] have shown that abnormalities in the Hippo pathway were closely related to the occurrence and development of colorectal cancer. In colorectal cancer and colorectal cancer-derived cell lines, YAP was overexpressed, and the activation of Mst1 and Mst2 protein kinases suppressed the overexpression of YAP, thus preventing the proliferation of intestinal stem cells and the occurrence of colorectal cancer [13]. However, studies on the effects of the regulation of Mst1 expression on the proliferation and apoptosis of colorectal cancer cells have not been reported. In this study, we performed the targeted silencing of Mst1 gene expression in SW480 colorectal cancer cells with an aim to investigate the impacts of regulation of Mst1 expression on the proliferation and apoptosis of colorectal cancer cells and to determine the underlying mechanisms. Our findings provide experimental evidence supporting the use of Mst1 in targeted therapy for colorectal cancer.
Materials and Methods
Cells and experimental grouping
Cells of the human colorectal cancer cell line SW480 (purchased from the Cell Bank of Type Culture Collection Committee, Chinese Academic of Sciences) were cultured in H-DMEM complete culture medium (GIBCO Co) containing 100 U/ml penicillin and 100 U/ml streptomycin at 37°C and 5% CO2. SW480 cells were collected when they were in the logarithmic growth phase and randomly divided into 7 groups:
1) Control group: the cells were seeded at a density of 2 × 105/well in 6-well plates, the culture medium was removed on the next day, and serum-free culture performed for another 6 h, after which the medium was replaced with complete medium for another 48 h of culture.
2) Group p-EGFP-N1: cells were transfected with the p- EGFP-N1 plasmid and cultured for 48 h.
3) Group p-EGFP-Mst1: cells were transfected with the p- EGFP-Mst1 plasmid and cultured for 48 h.
4) Group 20 μmol/l 5-FU: 20 μmol/L 5-FU (Ameresco Co) was added on the next day of seeding and the cells were cultured for 48 h.
5) Group 20 μmol/l 5-FU+p-EGFP-Mst1: cells were transfected with the p-EGFP-Mst1 plasmid on the next day of seeding and cultured for 24 h; 20 μmol/l 5-FU was then added and the cells were cultured for another 24 h.
6) Group NC-siRNA: cells were transfected with NC-siRNA and cultured for 48 h.
7) Group Mst1-siRNA3: cells were transfected with Mst1- siRNA3 and cultured for 48 h.
The transfection efficiency was determined after every transfection. The MTT assay was used for the determination of cell proliferation in each group, flow cytometry was used for determining the apoptosis rate and RT-qPCR and western blot analyses were performed to determine the gene expression in each group.
Transfection
PolyJetTM in vitro DNA transfection reagent was used to mediate the transfection of pEGFP-N1-Mst1 recombinant plasmid (Cat# SL100688; SigmaGen, Rockville, USA) and the empty vector plasmid pEGFP-N1 (Clontech, Mountain View, USA) into the SW480 cells. First, the SW480 cells were cultured in six-well plates until the cell confluence reached 60-70%; the medium was discarded before transfection, and 900 μl of fresh complete medium was added into each well, followed by incubation for 30-60 min. We then added 100 μl of transfection solution into each well, mixed the wells gently, and cultured them at 37°C and 5% CO2 for 5 h; 10% FBS-containing DMEM culture (TransGen Biotech, Beijing, China) was then added, and the cells were cultured for another 24-48 h. Fluorescence microscopy was then performed to observe and photograph the transfection efficiencies.
The transfection solution was prepared as follows: solution A was prepared by dissolving 1 μg of plasmid in 50 μl of serum-free DMEM medium; the solution was mixed by gentle repeated pipetting for 3-4 times. Solution B contained 3 μl of PolyJetTM reagent dissolved in 50 μl of serum-free DMEM medium, mixed by gentle repeated pipetting 3 to 4 times. Solution C (transfection solution) was prepared by quickly adding solution B into solution A followed by quick repeated pipetting 3 to 4 times. The solution was then allowed to stand for 15 min at room temperature.
Design and screening of siRNA
According to the human Mst1 (cDNA) sequence, the siRNA molecule was designed and synthesized by Shanghai Novobio Biological Technology Co., Ltd. The siRNA was double-stranded with a 21-bp sense strand including 19 bp of target sequence and 2 TT residues at the 3’ end, and a 21 bp antisense strand exactly complementary to the target sequence; the average molecular weight was 13,300. The sequences were identified to have no homology with any of the genes except Mst1 based on the sequence data obtained from GenBank. An appropriate amount of DEPC water was added to achieve a final concentration of 20 μM. Cells were seeded in six-well plates and cultured with FBS and antibiotic-containing DMEM medium (the number of inoculated cells should reach 50-70% confluence within 24 h). After the cells adhered to the walls of the plates after 4 h, appropriate amounts of the transfection reagent Lipofectamine TM2000 at a ratio of 1:1 were added to the siRNA for transfection.
MTT assay
The cells were collected and added to H-DMEM complete medium to obtain a single cell suspension. The cells were then seeded at a concentration of 1 × 103 to 1 × 104 cells/well into 96-well plates, with 200 μl of suspension in each well (the edges of the well were filled with sterile PBS). Transfection was performed and different concentrations of the drug were added for treatment. Each treatment was performed on three wells. After culturing in a cell incubator for 24 h, 48 h, or 72 h, 20 μl of MTT (Beyotime, Haimen, China) was added into each well, and the cells were cultured for another 4 h before the treatment was terminated. The wells were shaken for 10 min to fully dissolve the crystals. The absorbance in each well was detected at 490 nm 3-5 times, and the results were recorded. The rate of inhibition of cell proliferation was as determined as follows: Cell proliferation inhibition rate (%)=1-cell survival rate=(1-Absorbance of the experimental group at 490 nm/ Absorbance of the control group at 490 nm) × 100.
Flow cytometry
The SW480 cells were plated at a density of 1 × 105 cells/ml. After treatment for 48 h, 1-5 × 105 cells were collected and 500 μl of Binding Buffer was added to suspend the cells. 5 μl of Annexin V-FITC (MultiSciences, Hangzhou, China) was then added to the cell suspensions and mixed well. 5 μl of propidium iodide was added and mixed well. The reaction was allowed to occur in darkness and at room temperature for 5-15 min, followed by the detection of cell apoptosis by flow cytometry (Becton Dickinson, Facscallbar type), each analysis was performed on at least 30,000 events.
RT-qPCR
The primers for RT-qPCR were selected from http://pga.mgh.harvard.edu/primerbank/ and synthesized by GeneScript Biotechnology Co., with the primer sequences shown in Table 1. β-Actin was used as the internal standard, and the experiment was performed using SYBR®Premix Ex TaqTM II (Tli RNase H Plus) according to the manufacturer’s instructions. PCR was performed 40 cycles of: 95°C for 40 s, 58°C for 40 s, and 72°C for 60 s. The relative amount of target mRNA was determined using the relative expression values by normalizing the target mRNA Ct values to those for β-Actin (ΔCt).
| SiRNA sample | SiRNA sequence (5' to 3') | |
|---|---|---|
| Mst1-siRNA-245 | Sense | CCGGCCAGAUUGUUGCUAUTT |
| (Mst1-siRNA1) | Anti-sense | AUAGCAACAAUCUGGCCGGTT |
| Mst1-siRNA-716 | Sense | CCCUGGGAAUAACUGCCAUTT |
| (Mst1-siRNA2) | Anti-sense | AUGGCAGUUAUUCCCAGGGTT |
| Mst1-siRNA-1127 | Sense | CCAUGACUGAUGGAGCCAATT |
| (Mst1-siRNA3) | Anti-sense | UUGGCUCCAUCAGUCAUGGTT |
| Negative Control | Sense | UUCUCCGAACGUGUCACGUTT |
| (NC-siRNA) | Anti-sense | ACGUGACACGUUCGGAGAATT |
| Negative Control | Sense | UUCUCCGAACGUGUCACGUTT |
| FAM | Anti-sense | ACGUGACACGUUCGGAGAATT |
Table 1. Sequence of synthetic siRNA.
Western blot
Total proteins were extracted from the collected cells, followed by protein denaturation and SDS-PAGE. The total protein content of each well was 30 μg; the wet transferring method was used to transfer the proteins onto a Polyvinylidene Fluoride membrane (PVDF). Bands were detected immunologically using antibodies against PUMA (Cat. #55120-1-AP, Proteintech Co., USA), P53 (Cat. #21086-1, SAB Co.), P73 (Cat. #ab40658, Abcam Co.), Mst1 monoclonal antibody (Cat. #ab51134, Abcam Co.), YAP (Cat. #4912, Cell Signaling Technology Co.), P-YAP (Cat. #ab76252, Abcam Co.) and Caspase-3 monoclonal antibody (Cat. #9665, Cell Signaling Technology Co.) overnight at 4°C and at a ratio of 1:200. The membrane was then cultured with horseradish peroxidase-labeled secondary antibody (1: 2000, ZDR-5307, Beijing Zhongshan Golden Bridge Co.) for 1.5 h. Finally, ECL immunoassay was performed in the dark. Image-Pro Plus6.0 gel image analysis software was used to scan the gray scale values of protein bands for semi-quantitative analysis using β- actin as the control (AF7018, Santa Cruz Co.).
Statistical analysis
SPSS 19.0 statistical software was used for the analysis of experimental data. The comparisons between the groups were performed using ANOVA (one-way ANOVA) or an unpaired t-test, and the pairwise comparison between the groups was performed using the q test, with P<0.05 considered to indicate statistical significance.
Results
Plasmid transfection efficiency
The SW480 cells transfected with p-EGFP-N1 and p-EGFP-Mst1 plasmids, in which about 80% of the cells exhibited green fluorescence under fluorescence microscopy (Figure S1), while the control group showed no fluorescence. In addition, the NC-FAM-siRNA group also showed green fluorescence, suggesting that the siRNA fragment was transferred into the SW480 cells.
Proliferation and apoptosis of the SW480 cells
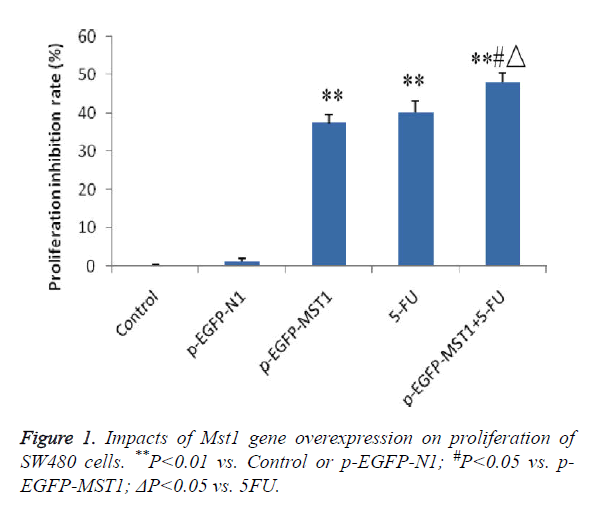
MTT colorimetric assay was performed to determine the effects of Mst1 gene overexpression on the proliferation of SW480 cells. The results showed that (Figure 1) the p-EGFP-Mst1, 5-FU and p-EGFP-Mst1+5-FU groups exhibited significantly higher growth inhibition rates than the control group or the p-EGFP-N1 group within 48 h (P<0.01). In addition, compared to the p-EGFP-Mst1 or 5-FU groups, the p- EGFP-Mst1+5-FU group exhibited significantly stronger proliferation inhibition (P<0.01), indicating that the overexpression of the Mst1 gene could inhibit the growth of SW480 cells, and the effects were enhanced with 5-FU.
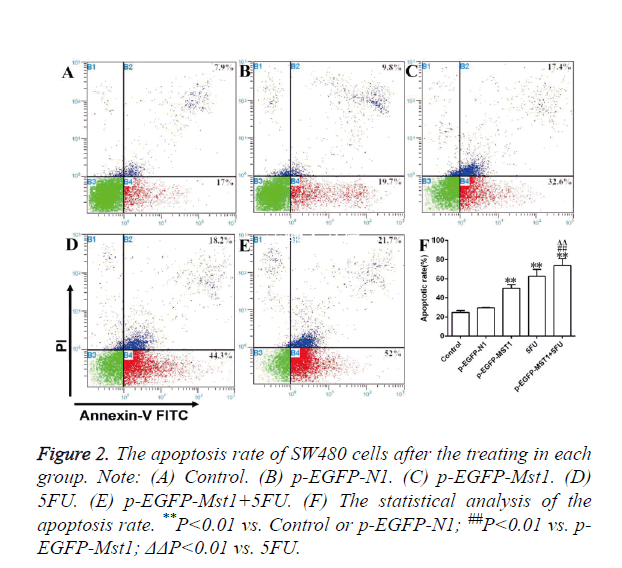
As determined by flow cytometry (Figure 2), the total apoptosis rate, early apoptosis rate (Annexin V-FITC+/PI-, right lower limit), and late apoptosis rate (Annexin V-FITC+/PI+, right upper limit) of the p-EGFP-Mst1 and 5-FU groups were significantly higher than those of the control and the p-EGFP-N1 groups (P<0.01). The total apoptosis rate and early apoptosis rate of the p-EGFP-Mst1+5FU group were significantly higher than those of the control and p-EGFP-N1 groups (P<0.05). The total apoptosis rate and early apoptosis rate of group p-EGFP-Mst1+5FU were also both significantly higher than those of the p-EGFP-Mst1 or 5FU groups (P<0.05), indicating that the Mst1 gene could promote the apoptosis of the SW480 cells; this effect would be further enhanced with the use of 5-FU.
mRNA expression
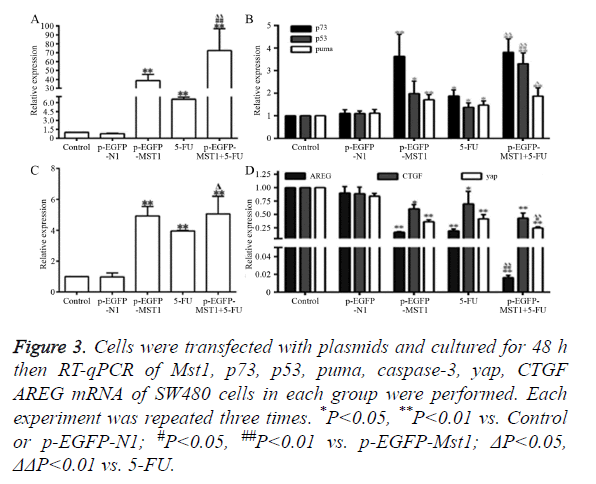
As shown in Figure 3, the mRNA expression of Mst1 in the p- EGFP-Mst1 and p-EGFP-Mst1+5FU groups was approximately 40-fold and 70-fold higher than that in the control group (Figure 3A); the expression in these groups was significantly higher than those in the control and p-EGFP-N1 groups (P<0.01). The mRNA expression of p73 (P<0.01), p53 (P<0.05), PUMA (P<0.01), and caspase-3 (P<0.01) in the p- EGFP-Mst1 group were increased by varying degrees. The expression of p73 increased by ~3.5-fold, that of p53 increased by ~2-fold, that of PUMA increased by ~1.8-fold, and that of caspase-3 increased by ~5-fold in comparison with the control and p-EGFP-N1 groups (Figures 3B and 3C). The mRNA expression of CTGF, AREG, and YAP in the p-EGFP-Mst1 group decreased to 0.2-, 0.6-, 0.4-fold of those in the control group (Figure 3D), and the differences in comparison to the expression in the control and p-EGFP-N1 groups were significant (P<0.05). The mRNA expression of CTGF, AREG, and YAP in the p-EGFP-Mst1+5FU group was decreased to about 0.2-fold of that in the control group, and the expression was significantly different from that in the p-EGFP-Mst1 (P<0.05) and 5FU (P<0.05) groups. This indicated that the overexpression of the Mst1 gene could upregulates the mRNA levels of p73, p53, PUMA, and caspase-3, while downregulating those of CTGF, AREG, and YAP.
Figure 3. Cells were transfected with plasmids and cultured for 48 h then RT-qPCR of Mst1, p73, p53, puma, caspase-3, yap, CTGF AREG mRNA of SW480 cells in each group were performed. Each experiment was repeated three times. *P<0.05, **P<0.01 vs. Control or p-EGFP-N1; #P<0.05, ##P<0.01 vs. p-EGFP-Mst1; ΔP<0.05, ΔΔP<0.01 vs. 5-FU.
Protein expression
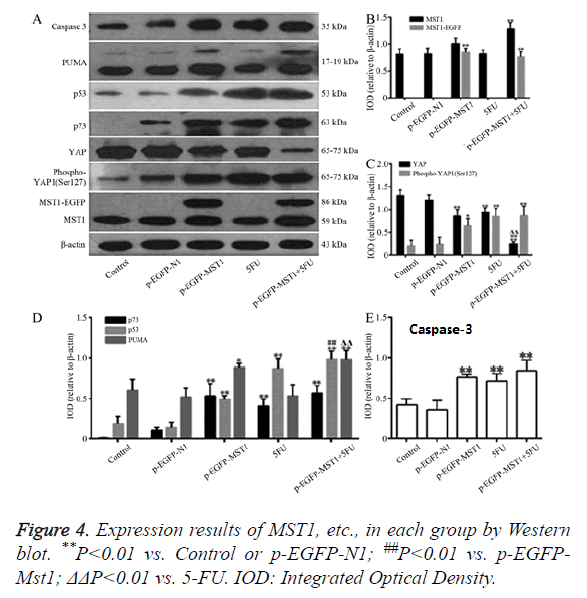
Western blot was used to determine the protein expression of Mst1, YAP, Phospho-YAP1 (Ser127) P73, P53, PUMA, and Caspase-3. As shown in Figure 4, compared to the control and p-EGFP-N1 groups, the total Mst1 protein in the p-EGFP-Mst1 group was increased by about 2.2-fold, and that of Phospho- YAP1 (Ser127)/YAP was increased by about 4.5-fold. The protein expressions of p73 (P<0.01), p53 (P<0.01), PUMA (P<0.05), and Caspase-3 (P<0.01) were significantly different from those in the control and p-EGFP-N1 groups. Total Mst1 protein expression in the p-EGFP-Mst1+5-FU group increased by about 2.5-fold when compared to that in the control and p- EGFP-N1 groups, and the expression of Phospho-YAP1 (Ser127)/YAP also increased by about 20-fold (Figure 4C). The protein expression of p73, p53, PUMA, and Caspase-3 exhibited significant differences compared to the expression in the control and p-EGFP-N1 groups (P<0.01). Compared with the pEGFP-Mst1 or 5-FU groups, the p-EGFP-Mst1+5-FU group showed a 4.5-fold increase in the expression of Phospho- YAP1 (Ser127)/YAP. The results showed that Mst1 overexpression could promote the phosphorylation of YAP in SW480 cells, and could enhance the protein expression of p73, p53, PUMA, and Caspase-3. The addition of 5-FU could further increase the expression of these proteins in the SW480 cells.
Effects of silencing of the Mst1 gene on the proliferation and apoptosis of SW480 cells
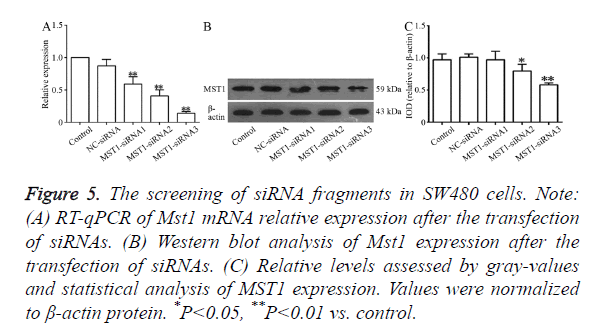
RT-qPCR (Figure 5A) showed that all the three siRNA fragments decreased Mst1 mRNA levels to varying levels. The differences in the Mst1 mRNA expression of the three siRNA fragments exhibited significant differences in comparison to the expressions in the control or NC-siRNA groups. The silencing efficiency in the Mst1-siRNA3 group was the highest (P<0.01) at about 85.7%. Western blot results, as shown in Figure 5B, indicated that the transfection with these three siRNA segments downregulated the expression of the Mst1 protein to varying degrees. The largest degree of downregulation was observed in the Mst1-siRNA3 group (0.580 ± 0.029) (about 41.1%), and the expression was significantly different from that in the control group (P<0.01).
Figure 5. The screening of siRNA fragments in SW480 cells. Note: (A) RT-qPCR of Mst1 mRNA relative expression after the transfection of siRNAs. (B) Western blot analysis of Mst1 expression after the transfection of siRNAs. (C) Relative levels assessed by gray-values and statistical analysis of MST1 expression. Values were normalized to β-actin protein. *P<0.05, **P<0.01 vs. control.
Based on these results, we selected the Mst1-siRNA3 fragment as the fragment for further use in the targeted silencing of Mst1.
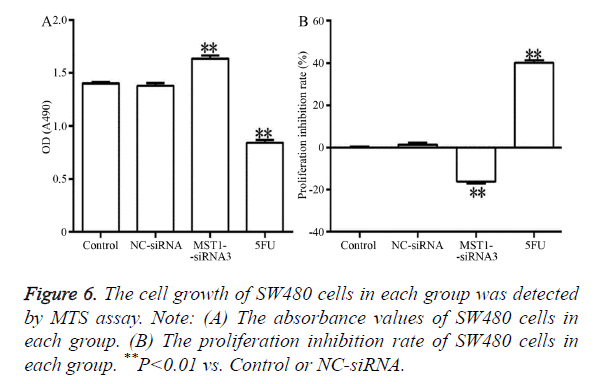
Figure 6 showed that, after 48 h of transfection, the absorbance in the Mst1-siRNA3 group was significantly higher than that in the control and NC-siRNA groups (Figure 6A) (P<0.01). In addition, the proliferation rate in this group was negative, indicating that cell proliferation was promoted (Figure 6B). The differences in comparison to the control and NC-siRNA groups were statistically significant (P<0.01).
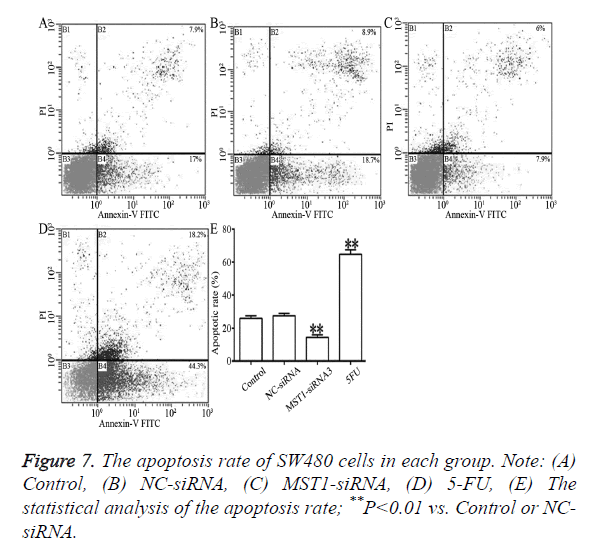
As shown in Figure 7, compared to the control and NC-siRNA groups, the Mst1-siRNA3 showed significantly lower rates of total apoptosis, early apoptosis (Annexin V-FITC+/PI-, right lower limit), and late apoptosis (Annexin V-FITC+/PI+, upper limit) (P<0.01), which indicated that Mst1 silencing could inhibit the apoptosis of the SW480 cells.
Effects of the targeted silencing of the Mst1 gene on the mRNA expression of p73, p53, YAP, PUMA, CTGF, AREGE, and caspase-3
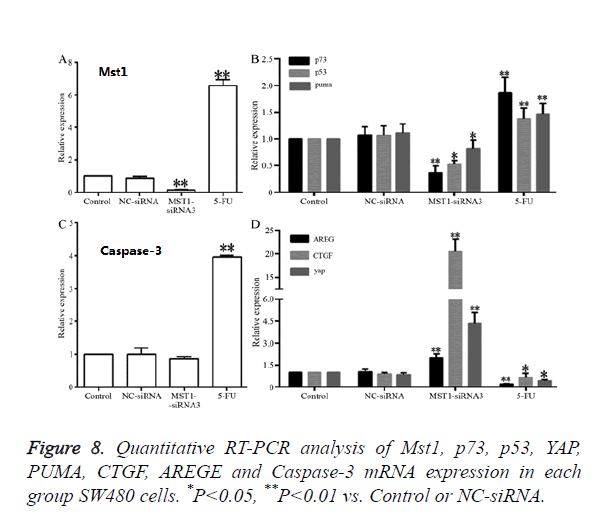
RT-qPCR results revealed that (Figure 8), compared to the control and NC-siRNA groups, the Mst1-siRNA3 group showed a significantly lower Mst1 mRNA expression (Figure 8A); the Mst1 mRNA expression was about 0.15-fold of the control group, and the difference was statistically significant (P<0.01). The mRNA expression of p73 (P<0.01), p53 (P<0.05) and PUMA (P<0.05) was also decreased by different degrees, and the differences were significant in comparison to those in the control and NC-siRNA groups (Figures 8B and 8C). In addition, the mRNA expression of CTGF, AREG, and YAP in the Mst1-siRNA3 group were 20-, 2-, and 4-fold higher than those in the control group (Figure 8D), and the differences were statistically significant (P<0.01), indicating that Mst1 silencing could downregulates the mRNA levels of p73, p53, and PUMA, while upregulating the mRNA levels of CTGF, AREG, and YAP.
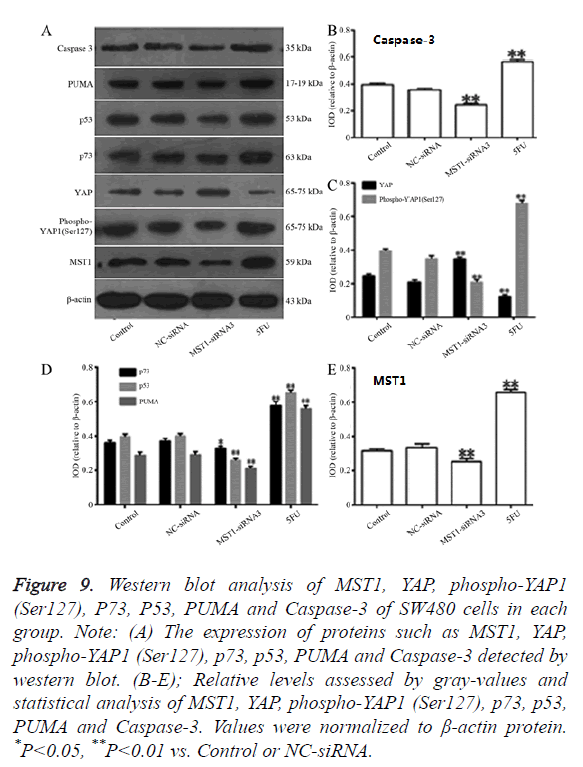
Effects of the targeted silencing of the Mst1 gene on the protein expression of YAP, P73, p53, PUMA, and Caspase-3 and the phosphorylation level of YAP
Western blot assay revealed that (Figure 9) the MST1 protein expression in the Mst1-siRNA3 group was 0.6-fold of the control group (Figure 9B), and the ratio of Phospho-YAP1 (Ser127)/YAP was 0.3-fold of that in the control group (Figure 9C). The protein expression of P73 (P<0.05), P53 (P<0.01), PUMA (P<0.01), and Caspase-3 (P<0.01) was significantly lower than that in the control or NC-siRNA groups (Figures 9D and 9E), indicating that Mst1 silencing could downregulate the protein expression of p73, p53, PUMA, and Caspase-3 in the SW480 cells, and inhibit the phosphorylation of the YAP protein.
Figure 9. Western blot analysis of MST1, YAP, phospho-YAP1 (Ser127), P73, P53, PUMA and Caspase-3 of SW480 cells in each group. Note: (A) The expression of proteins such as MST1, YAP, phospho-YAP1 (Ser127), p73, p53, PUMA and Caspase-3 detected by western blot. (B-E); Relative levels assessed by gray-values and statistical analysis of MST1, YAP, phospho-YAP1 (Ser127), p73, p53, PUMA and Caspase-3. Values were normalized to β-actin protein. *P<0.05, **P<0.01 vs. Control or NC-siRNA.
Discussion
Colorectal cancer is one of the most common human malignancies, with its incidence rate ranked third among the malignant tumors in the world [19]. Studies have shown that Mst1 could be a potential therapeutic target for tumor [1,8] or chronic vascular diseases. Xu et al. [1] found that when Mst1 was transfected into Non-Small Cell Lung Cancer (NSCLC) cells, Mst1 would be re-expressed, thus inactivating the oncogene Yap1 and phosphorylating Yap1 at the Ser127 residue, which eliminated the tumorigenicity and promoted apoptosis of the NSCLC cells. Ono et al. [20] introduced Ad- Mst1 into the vascular smooth muscle cells, and found the number of apoptotic cells to increase. This indicated that Mst1 could promote the apoptosis of vascular smooth muscle cells. The Mst1 gene in the Hippo signaling pathway can inhibit the accumulation of the YAP protein in cell nuclei, thus limiting cell growth; it was also found that the expression of the YAP protein in colorectal cancer was the highest [14]. However, the cell proliferation and tumorigenesis caused by the loss of the Mst1 gene in Drosophila and murine animals could be neutralized by the reduction or absence of YAP, suggesting that the Mst1 gene might control cell proliferation and tumorigenesis through YAP [21]. The results of this study showed that overexpression of Mst1 increased the ratio of Phospho-YAP1 (Ser127)/YAP as well as the phosphorylation level of YAP. Further, the expression of apoptosis-related genes, such as p73, p53, PUMA, and caspase-3, was increased, while the mRNA levels of downstream transcription factors downstream of YAP, as CTGF and AREG, were correspondingly decreased. Cell proliferation was reduced while apoptosis was increased.
When Mst1 was silenced, the ratio of Phospho-YAP1 (Ser127)/YAP decreased, YAP was less phosphorylated, the expression of p73, p53, PUMA, and Caspase-3 reduced, and the mRNA levels of CTGF and AREG correspondingly increased, so that the proliferation rate of the SW480 cells increased while the apoptosis rate decreased. These results confirmed that the Mst1 gene could control cell proliferation, apoptosis, and tumorigenesis through YAP. Strano et al. [22,23] found that after YAP was phosphorylated to Phospho-YAP, without treatment with CDDP, Phospho-YAP (Ser127) would combine with the 14-3-3 protein inside the cytoplasm. However, after treatment with CDDP, Phospho-YAP (Ser127) might dissociate from the 14-3-3 protein, enter nuclei, and then combine with PML, p73, and p300 to form a transcriptional activator complex. When apoptotic conditions are satisfied, the complex promotes the transcription of p53AIP1 (p53-regulated apoptosis-inducing protein 1) and Bax, leading to apoptosis. p53AIP1 is a target gene of p53, which can mediate p53- dependent apoptosis [24], and p53-dependent apoptotic target genes include Bax [25], Fas [26,27], and PUMA [28,29]. Therefore, we believe that the targeted regulation of Mst1 expression in cells can be used in the clinical treatment of colorectal cancer. Mst1 can be used to promote the phosphorylation of YAP (Ser127) inside the cytoplasm, so that the Phospho-YAP1 inside cytoplasm will dissociate from the 14-3-3 protein, enter nuclei, and act with p73 to increase the expression of p53 and PUMA, activate caspase-3, and enhance the apoptosis of the cancer cells. At the same time, after Mst1 expression is increased, the content of YAP entering the nuclei from the cytoplasm would be reduced, resulting in decrease in the expression of CTGF and AREG, thereby reducing the expressions of cell proliferation-related genes and inhibiting cell proliferation.
Acknowledgments
This study was supported by the National Natural Science Foundation of China No.81360032 (FSW), No. 81360313 (ZQL), the Natural Science Foundation of Jiangxi Province No. 20171BAB205055 (ZQL).
Conflicts of Interest
All of the authors declare that they have no conflicts of interest regarding this paper.
References
- Xu CM, Liu WW, Liu CJ, Wen C, Lu HF. Mst1 overexpression inhibited the growth of human non-small cell lung cancer in vitro and in vivo. Cancer Gene Ther 2013; 20: 453-460.
- Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development 2011; 138: 9-22.
- Song H, Mak KK, Topol L, Yun K, Hu J, Garrett L, Chen Y, Park O, Chang J, Simpson RM, Wang CY, Gao B, Jiang J, Yang Y. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc Natl Acad Sci USA 2010; 107: 1431-1436.
- Leberer E, Dignard D, Harcus D, Thomas DY, Whiteway M. The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein beta gamma subunits to downstream signalling components. EMBO J 1992; 11: 4815-4824.
- Goehring AS, Mitchell DA, Tong AH, Keniry ME, Boone C, Sprague GF Jr. Synthetic lethal analysis implicates Ste20p, a p21-activated potein kinase, in polarisome activation. Mol Biol Cell 2003; 14: 1501-1516.
- Ahn SH, Cheung WL, Hsu JY, Diaz RL, Smith MM, Allis CD. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell 2005; 120: 25-36.
- Ahn SH1, Henderson KA, Keeney S, Allis CD. H2B (Ser10) phosphorylation is induced during apoptosis and meiosis in S. cerevisiae. Cell Cycle 2005; 4: 780-783.
- Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, Lauwers GY, Thasler W, Lee JT, Avruch J, Bardeesy N. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell 2009; 16: 425-438.
- Dan I, Watanabe NM, Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol 2001; 11: 220-230.
- Lu L, Li Y, Kim SM, Bossuyt W, Liu P. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci USA 2010; 107: 1437-1442.
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 2005; 122: 421-434.
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 2007; 130: 1120-1133.
- Zhou D, Zhang Y, Wu H, Barry E, Yin Y, Lawrence E, Dawson D, Willis JE, Markowitz SD, Camargo FD, Avruch J. Mst1 and Mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis by inhibition of Yes-associated protein (Yap) overabundance. Proc Natl Acad Sci USA 2011; 108: 1312-1320.
- Konsavage WM, Kyler SL, Rennoll SA, Jin G, Yochum GS. Wnt/beta-catenin signaling regulates Yes-associated protein (YAP) gene expression in colorectal carcinoma cells. J Biol Chem 2012; 287: 11730-11739.
- Xu C, Liu C, Huang W, Tu S, Wan F. Effect of Mst1 overexpression on the growth of human hepatocellular carcinoma HepG2 cells and the sensitivity to cisplatin in vitro. Acta Biochim Biophys Sin 2013; 45: 268-279.
- Zhou GX, Li XY, Zhang Q, Zhao K, Zhang CP. Effects of the hippo signaling pathway in human gastric cancer. Asian Pac J Cancer Prev 2013; 14: 5199-5205.
- Liang K, Zhou G, Zhang Q, Li J, Zhang C. Expression of hippo pathway in colorectal cancer. Saudi J Gastroenterol 2014; 20: 188-194.
- Tschaharganeh DF, Chen X, Latzko P, Malz M, Gaida MM, Felix K, Ladu S, Singer S, Pinna F, Gretz N, Sticht C, Tomasi ML, Delogu S, Evert M, Fan B, Ribback S, Jiang L, Brozzetti S, Bergmann F, Dombrowski F, Schirmacher P, Calvisi DF, Breuhahn K. Yes-Associated Protein Up-regulates Jagged-1 and Activates the NOTCH Pathway in Human Hepatocellular Carcinoma. Gastroenterology 2013; 144: 1530-1542.
- Suo T, Mahteme H, Qin XY. Hyperthermic intraperitoneal chemotherapy for gastric and colorectal cancer in Mainland China. World J Gastroenterol 2011; 17: 1071-1075.
- Ono H, Ichiki T, Ohtsubo H, Fukuyama K, Imayama I. Critical role of Mst1 in vascular remodeling after injury. Arterioscler Thromb Vasc Biol 2005; 25: 1871-1876.
- Qin F, Tian J, Zhou D, Chen L. Mst1 and Mst2 kinases: regulations and diseases. Cell Biosci 2013; 3: 31.
- Strano S, Monti O, Pediconi N, Baccarini A, Fontemaggi G, Lapi E, Mantovani F, Damalas A, Citro G, Sacchi A, Del Sal G, Levrero M, Blandino G. The transcriptional coactivator Yes-associated protein drives p73 gene-target specificity in response to DNA Damage. Mol Cell 2005; 18: 447-459.
- Basu S, Totty NF, Irwin MS, Sudol M, Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol Cell 2003; 11: 11-23.
- Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 2000; 102: 849-862.
- Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 1995; 80: 293-299.
- Owen-Schaub LB, Zhang W, Cusack JC, Angelo LS, Santee SM, Fujiwara T, Roth JA, Deisseroth AB, Zhang WW, Kruzel E. Wild-Type human P53 and a temperature-sensitive mutant induce Fas/Apo-1 expression. Mol Cell Biol 1995; 15: 3032-3040.
- Müller M, Wilder S, Bannasch D, Israeli D, Lehlbach K. p53 activates the CD95 (APO-1/Fas) gene in response to DNA damage by anticancer drugs. J Exp Med 1998; 188: 2033-2045.
- Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell 2001; 7: 683-694.
- Tu S, Zhang X, Luo D, Liu Z, Yang X. Effect of taurine on the proliferation and apoptosis of human hepatocellular carcinoma HepG2 cells. Exp Ther Med 2015; 10: 193-200.