ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 6
CTLA-4 gene polymorphism A/G in exon 1 (+49) is associated with plasma viral load in HIV-1 infection patients
Wenjuan Tang1#, Fang Xie2#, Xiaoli Zeng1#, Hong Sun3# and Haibo Zhao4*
1Department of Maternal Health Care, Shiyan Maternal and Child Health Hospital, Shiyan, Hubei, PR China
2Department of Gynaecology, Shiyan Maternal and Child Health Hospital, Shiyan, Hubei, PR China
3Department of Geriatrics, WuHan hospital of Chinese traditional and western medicine, WuHan, Hubei, PR China
4Department of Paediatric Surgery, Shiyan Maternal and Child Health Hospital, Shiyan, Hubei, PR China
#These authors contributed equally to this work
- *Corresponding Author:
- Haibo Zhao
Department of Paediatric Surgery
Shiyan Maternal and Child Health Hospital, PR China
Accepted on November 4, 2016
Objective: Cytotoxic T Lymphocyte Antigen 4 (CTLA-4) is a potent immunoregulatory molecule that suppresses antitumor response by down-regulating T cell activation and is believed to have a critical modulatory role in the immune response. The expression and function of CTLA-4 may be affected by gene polymorphisms. The present study examined the correlation of CTLA-4 gene +49 G/A polymorphism with HIV-1 infection, and evaluated the role of +49G/A polymorphism on plasma EFV level and treatment response in the form of change in HIV viral load, CD+4 T-cell counts and CD+4/CD +8 ratios in HIV-1 infected patients receiving Highly Active Antiretroviral Therapy (HAART).
Materials and Methods: The CTLA-4 exon 1 +49 A/G Single Nucleotide Polymorphism (SNPs) were genotyped in 45 adult patients with HIV-1 infection and 50 ethnically, age, gender matched healthy controls through polymerase chain reaction. Plasma EFV levels were detected using high-performance liquid chromatography, and viral load, CD+4 T-cell counts and CD+4/CD+8 ratios were measured at enrolment and every 2 months for 6 months.
Results: Frequencies of the CTLA-4 +49 AG and GG genotype and the +49 G allele were not significantly increased in patients with HIV-1 infection compared to healthy controls (AG andGG vs. AA: Odds Ratio (OR)=1.70, 95% confidence interval (CI): 0.74-3.89, P=0.21; G vs. A: OR=1.45, 95% CI: 0.74-2.82, P=0.28), meanwhile, were not significantly associated with high plasma EFV concentrations (P=0.0236), whereas the GG genotype was associated with higher plasma viral load (P=0.017) and lower CD+4 T-cell counts (P=0.009). The relationship between the reactivation of HIV viral load declines, increased CD+4 T-cell counts and increment ratio of CD+4/CD+8 in peripheral blood were observed.
Conclusions: This study provides the first evidence that CTLA-4 exon 1 +49A/G polymorphism does not represent a major susceptibility risk allele for HIV-1 infection, however, supporting the association with plasma HIV viral load level. Furthermore, the CTLA-4 (+99A/G) gene polymorphism is not associated with immune response to HAART. To validate our results, further studies on a larger cohort are needed.
Keywords
CTLA-4, Polymorphism, HIV-1 infection, Viral Load, Immune response.
Introduction
Human Immunodeficiency Virus (HIV) causes HIV infection and Acquired Immune Deficiency Syndrome (AIDS). Globally, statistics from the WHO and UNAIDS showed that approximately 2.3 million people became newly infected and 1.6 million died of AIDS-related causes at the end of 2012, meanwhile, there are up to more than 40 million people currently living with HIV infection worldwide [1]. In China in 2014, an estimated 0.448 million Chinese are living with HIV/ AIDS and 0.139 million deaths were registered [2]. HIV continues to be a major global public health issue, and there is no cure for HIV infection by far. Infection with HIV type 1 (HIV-1) typically occurs across mucosal surfaces or by direct inoculation. Once HIV-1 infects a cell, the virus integrates into host genetic material and either begins cycles of replication or remains inactive, causing latent infection in cellular reservoirs [3]. Emerging evidence suggests that early events in HIV-1 infection may play a critical role in determining disease progression and environmental and genetic factors play an important role in the development of disease [4]. The inhibitory immunoregulatory molecule Cytotoxic T Lymphocyte Antigen (CTLA)-4 has been associated with HIV-1 control, and the programmed death-1 molecule with CD +4 T-cell exhaustion [5,6]. CTLA-4, namely, CD152, encoded by a gene on chromosome 2q33, is a receptor mainly expressed by activated T lymphocytes and may down-regulate the second signal to T-cells [7,8]. As an important regulator, CTLA-4 functions negatively in immune response and counteracts the stimulation initiated by the CD28 molecule [9,10]. Since the CTLA-4 gene product has inhibitory effects on the immune system, any variation in its expression or function may lead to the breakdown of the delicate homeostasis of this system. Indeed, several researches have confirmed that CTLA-4 gene polymorphisms are implicated in both autoimmune diseases and malignancy susceptibility [11-15]. Moreover, mice deficient in CTLA-4 may develop severe autoimmune disease and lymphoproliferative disorder [16,17], and some certain gene positions are linked to severe virus-associated autoimmune diseases, the position 49 A/G of exon 1 (dbSNP: rs231775) is largely associated, e.g. Enterovirus 71 [18], human T-cell lymphotrophic virus-1 [19], HCV [13], HBV [20,21], together with some other virus-associated diseases, which have been validated in some previous studies. Therefore, is there some relationship between the position 49 A/G of CTLA-4 exon 1 polymorphism and the risk of HIV-associated disease? As seldom are there studies done for the relationship between 49 A/G of CTLA-4 gene polymorphisms and the risk of HIV/AIDS, and given the pivotal role of CTLA-4 in regulating the immune response, we designed this study to test the hypothesis that specific CTLA-4 SNPs (i.e.t49 A/G) are associated with risk of the HIV-1 infection. Plasma HIV Viral Load (PHVL) and CD+4 T-cell counts are two surrogate markers of HIV-1 disease progression. Studies have shown that T cells in patients with high PHVL and progressive disease are less functional, have less proliferative capacity, and are more exhausted than T cells in patients with low PVL and slow disease progression [22,23]. HIV-1 induces a sustained inflammatory response with a large proportion of CD+4 T-cell counts expressing cell surface activation markers [24]. It has been showed that CTLA-4 is up regulated on HIV-specific CD +4, but not CD+8, T cells and in vitro blockade of CTLA-4 augment HIV-specific CD+4 T-cell function [6,7]. Up regulation of CTLA-4 increases CCR5 expression and enhances susceptibility of CD+4 T-cell to HIV infection [25,26]. Consequently, it is supposed that CTLA-4 gene polymorphism expression may correlate with markers of susceptibility of HIV disease. Drug treatment in HIV disease is characterized by variable responses, in terms of both efficacy and toxicity. Genetic and environmental factors are important determinants of this variability, although the relative contributions are unclear and are likely to vary with different drugs [27]. Efavirenz (EFV) is a non-nucleoside reverse transcriptase inhibitor that has demonstrated appropriate efficacy and safety in the treatment of HIV-1 infection in many clinical studies [28,29]. It is therefore often used in Highly Active Anti- Retroviral Therapy (HAART) implemented for the treatment of both native and experienced patients. Several studies have confirmed that genetic differences among individuals influence metabolism, distribution and elimination of EFV [30,31], however, the evidence that genotyping and measurement of EFV plasma concentrations actually improve patient outcome is lacking. Plasma HIV virus load and CD+4 T-cell counts are two important biomarkers for assessing clinical course of HIVinfected patients [22,32], predicting clinical progress and evaluating therapy of anti-HIV drugs. The objectives of this investigation were to explore the association between CTLA-4 exon 1 polymorphism (49 A/G) and plasma EFV concentration, clinical response of EFV as well.
Patients and Blood Samples
Patient selection
The study was conducted at the Taihe hospital in Shiyan of HuBei province, and patients, recruited from the whole hospital outpatients aged 16-50 years were to the amount of 45. Primary HIV-1 infection was diagnosed as before by a positive Enzyme-Linked Immunosorbent Assay (ELISA) result in hospital, moreover, it was confirmed via Western Blotting (WB). Before entering the study, the medical histories of the patients were recorded, physical examination and haematological, biochemical and virological tests were performed. All of them had not previously been treated with lamivudine or any non-nucleoside reverse-transcriptase inhibitor or protease inhibitor, and additional exclusion criteria included recent receipt of a vaccine or blood transfusion, other cause of fever, evidence of underlying chronic diseases such as infection with Hepatitis virus, diabetes, impaired kidney function, cardiac-cerebral vascular disease including high blood pressure or coronary heart disease and any other immune system disease. All Male and female outpatients were eligible for study enrolment except pregnant women. Although this was not a randomized trial, 50 HIV-1-seronegative subjects served as controls, the control volunteers were identified on the basis of the following criteria: (a) diagnosis of no infection with the Hepatitis virus of A, B, C, D or E; (b) no history with the diseases of liver, kidney or blood; (c) without any immune system disease or cardiac-cerebral vascular disease. Approval for the study was obtained from the local Ethics Committee and each patient provided written informed consent after appropriate review, discussion and counselling by the clinical study team. Blood Samples HIV-1 infected patients were assigned to receive the following combination of fixed-dose drugs orally daily within a 24-week follow-up: EFV (600 mg/ day, at bed time) plus zidovudine (AZT) (300 mg bid twice daily, 12 hours apart) and lamivudine (3TC) (150 mg twice daily), but not recommended to modify their daily activities and no specific dietary restrictions were given. As the fixed dose AZT/3TC tablet is not suitable for people weighing less than 50 kg, health care workers had been on the lookout for the relevant adverse effects and palliative therapy have been managed and provided. Meanwhile, HIV-1 infected patients were followed with a clinical assessment and routine laboratory monitoring every 8 weeks until week 24. Two tubes (2 ml per tube) of peripheral blood specimens per person were collected in EDTA liquid anticoagulant Vacutainer Tubes (Becton-Dickinson) from elbow vein during the morning time before eating breakfast. One of them was stored at 4°C for evaluating the absolute number of CD+4 and CD+8 T-cell counts using flow cytometry, the other were immediately centrifuged and the separated plasma was stored at -80°C before it was used for virus analysis using High Performance Liquid Chromatography (HPLC) and extracted peripheral lymphocytes for genomic DNA. Furthermore, individual patient information of epidemiology and larithmics was carefully recorded during follow-up visits, including dose history, sampling time, sex, age, weight, height and so on.
Study Design and Methods
Cytometry analysis flow cytometry was carried out with a Becton Dickinson FACScan. The cells were first stained in TruCount tube which contains CaliBRITEAPC. Blood samples were pipetted into the tube; 50 μl internal standard and 25 μl Multi-Test CD3-FITC/CD8-PE/CD45 PerCP/CD4 APC antibody (Becton Dickinson) were added. The contents were mixed thoroughly and incubated in dark place for 15 minutes at room temperature, and then added 450 μL FACS haemolysin into the tube (Becton Dickinson) and the operational approach was the same as described above. After staining, samples were injected and four-colour flow cytometric analysis was performed using FACSCalibur for the absolute cell count (cells per microliter) of CD+4 and CD+8 T-cells. The flow cytometry system used for this study was the CaliBRITETM, FACSCompTM and MultiSETTM software, which can analysis the result automatically. Polymorphism at position +49 of exon 1 of the CTLA-4 gene genomic DNA was isolated from whole blood samples by using a genomic DNA Minipreps Kit (Sangon, China) and used for Polymerase Chain Reaction (PCR). Genomic DNA was purified by use of the QIAamp blood reagent set (Qiagen) and assessed quantity and purity on the basis of spectroscopic absorbance at 260 and 280 nm. A fragment of the CTLA-4 gene was amplified with the following oligonucleotides: forward 5'-GCT CTA CTT CCT GAA GAC CT-3' and reverse 5'-AGT CTC ACT CAC CTT TGC AG-3', according to the sequence of human cDNA of CTLA-4 described by Kucharska et al. PCR was performed using genomic DNA (0.2 mg), Taq polymerase (1 U), 10 pmol of each primer, and dNTPs (200 mM) under the following conditions: initial denaturation for 5 min at 94°C, annealing for 45 s at 57°C, extension for 45 s at 72°C, denaturation for 45 s at 94°C (30 cycles), and a final extension for 8 min at 72°C. The presence of G alleles was determined in each subject by PCR amplification of CTLA-4, followed by digestion with restriction enzyme Bbv1 (New England BioLabs, Beverly, MA), which acts on the G variation, but not on the A variation. If a G allele was at position 49, 88/74-bp fragments were obtained. PCR products were detected by electrophoresis in a 3.5% NuSiev GTG agarose (FMC BioProducts, Rockland, ME) gels.
Viral-load assays Quantifying HIV Load Viral RNA copy numbers were determined by Reverse Transcriptase Polymerase Chain Reaction (RT-PCR) using the Amplicor HIV Monitor Kit (Roche Molecular Systems, Branchburg, NJ) according to the manufacturer’s instructions. HIV RNA was extracted from plasma stored previously with Cobas TaqMan reagent and then amplified, during all extractions and purifications, precautions were taken to reduce the risk of false-positive results. In brief, the assay uses HIV RT enzyme to catalyse the conversion of RNA to cDNA. For the first round of amplification, a forward primer was selected from the conserved Alu repetitive element of the human genome and the reverse primer represented the conserved gag-pol region of the HIV genome. The internal control plasmid contains the same primer binding regions as the target sequence and is of similar length and base composition but contains a unique internal probe binding region that allows it to be detected separately from the target sequences.
Amplification process was performed according to following profile: A 10 minute incubation at 50°C was followed by two cycles, with each cycle consisting of 20 s at 98°C, 20 s at 62°C, and 45 s at 72°C. This was followed by 41 cycles, with each cycle consisting of 20 s at 94°C, 20 s at 62°C, and 45 s at 72°C. A final incubation at 72°C for 5 min was included to allow for completion of strand synthesis. Due to the production of irregular signals from the real-time PCR, the latter samples were cleaned using the cleaning and elution steps of the QIAmp DNA Blood Mini kit (Qiagen), and eluted in 100 μl AE buffer. Subsequently, TaqMan PCR was used to determine the copy numbers of the pre-amplified HIV-1 RNA, specificity of the test was accomplished by hybridization of the amplified product, termed an amplicon. And emphasized necessarily, all PCR reactions were carried out with negative controls in a designated PCR clean room. Sample PHVL was compared against quantitation standard RNA, and calculated as HIV-1 RNA copies per millilitre of plasma. The lower and upper limit of detection for this assay are 4 × 10 copies/ml and 10 × 107 copies/ml, respectively.
Plasma EFV quantification Plasma EFV concentrations were measured by High-Performance Liquid Chromatography (HPLC) (Waters, Milford, MA) with UV detection at 215 nm, following solid-phase extraction using a GX-271 ASPEC (Gilson, Villiers le Bel, France). Briefly, an internal standard (20 μl, 500 ng/ml) (catalog no. Ro31-9564; Roche Discovery, Welwyn, United Kingdom) and acetonitrile (400 μl; VWR Laboratory Supplies, Poole, United Kingdom) were added to aliquots (100 μl) of calibrators, Quality Controls (QCs), and patient plasma. After mixing thoroughly and a liquid-liquid extraction for 10 min, the mixture was centrifuged for 5 min at 4000 g. Then the organic phase was transferred to clean tubes and evaporated to dryness. Subsequently, ammonium formate buffer (100 μl, 20 mM; Fisher Scientific, Loughborough, United Kingdom) were added into the clean tubes to reconstituted extracts into the HPLC mobile phase (0.1 ml) and transferred to vials for injection into the HPLC. The mobile phase was pumped at a flow rate of 1.2 ml/min and analysed by HPLC-tandem mass spectrometry (Waters Quattro Premier XE; Waters Corporation, MA) via an Acuity UPLC bridged ethyl hybrid C 18 column (Waters Corporation, MA). Ultraviolet detection was carried out at a wavelength of 247 nm. Retention times for EFV and Internal Standard (IS) were 5.3 and 4.5 minutes, respectively, and there was no chromatographic interference from other commonly administered drugs.
Statistical analysis
Allele and genotype frequencies were compared between groups using the Chi-square (χ2) test or Fisher’s exact probability test, where appropriate. Association analysis was performed by means of logistic regression. Genotype-based Odds Ratios (ORs) were calculated using individuals homozygous for the non-susceptible allele as the reference. Allele specific ORs were calculated under the assumption of a multiplicative risk model. ORs were calculated as a measurement of strength of association according to Woolf’s method. Quantitative results were expressed as mean ± Standard Deviation (SD). Student's t test is used to compare means of same variable between groups and the significance among three genotypes was assessed by Mann-Whitney U test. Statistical analysis was performed using SPSS statistical package 12.0 (SPSS Inc., Chicago, IL, USA). A difference at P<0.05 level was considered statistically significant.
Results
Baseline characteristics
This study was approved by the hospital ethics committee and all subjects gave the written informed consent to participate. Demographic and clinical characteristics are shown in Table 1. We recruited total 45 HIV-1 infected patients, including 25 (55.56%) were male and 20 (44.44%) were female, and ages ranged from 19-40 (28.44 ± 5.19) years. The control group consisted of 50 healthy subjects from two sources: hospital and community, matched by sex, age, ethnicity, education, residency area, socioeconomic status and smoking status. There were no significant differences in the distribution between cases and controls (P>0.05), suggesting that the matching based on these four variables was satisfactory. However, there was a significant difference in body weight, body mass index, CD+4 T-cell counts and CD+4?CD+8 ratios between the two groups (P ≤ 0.001). From the illustration of the risk factors associated with HIV-1 infection, it was the sexual transmission (35.56%) which was the most hazard factor for being infected. Meanwhile, the mean PHVL concentration in the HIV-1 group was 4.67 ± 0.64 (log 10 copies/ml). Meanwhile, the initial CD+4 T-cell counts and value of CD+4/CD+8 ratios in control group were higher than HIV-1 group by 489.79 ± 45.85, 0.99 ± 0.19 respectively, with significant difference (P<0.001), which inflected that the clinical characteristics of HIV-1 group and healthy group were different.
| Variable | Value (mean ± SD (95% CI) or no. (%)) | ||
|---|---|---|---|
| HIV-1 group | Healthy controls | t or χ2 (P-value) | |
| Gender (male/female) | 25/20 | 31\19 | χ2=0.41 (P=0.524) |
| Age (years) | 28.44 ± 5.19 (26.89?32.07) | 28.86 ± 6.21 (27.10?30.62) | t=0.35 (P=0.725) |
| Ethnicity | Han Chinese | Han Chinese | |
| Body weight (kg) | 57.36 ± 8.55 (54.78?59.92) | 63.52 ± 9.25 (60.89?66.15) | t=3.36 (P=0.001) |
| Body mass index (kg2/cm) | 20.05 ± 5.19 (18.45?21.56) | 24.10 ± 6.17 (22.34?25.85) | t=3.48(P=0.001) |
| Smoking patients | 29 (64.44%) | 33 (66.00%) | χ2=0.33 (P=0.564) |
| HIV risk factors | |||
| IV drug use | 4 (8.89%) | ||
| sexual transmission | 16 (35.56%) | ||
| Blood product transfusion | 7 (15.56%) | ||
| Maternal transmission | 4 (8.89%) | ||
| Uncertain | 14 (31.11%) | ||
| Initial CD+4 count, cells/mm3 | 384.24 ±38.69 (372.62?395.87) | 874.03 ± 84.54 (850.02?898.07) | 36.90 (p<0.001) |
| Initial valueof CDM+4?CD+8 ratio | 0.31 ± 0.07 (0.29?0.33) | 1.30 ± 0.26 (1.23?1.38) | 24.77 (p<0.001) |
| Initial HIV-1 RNA, log10 copies/ml | 4.67 ±0.64 (4.48?4.87) | ||
Table 1. Demographic and clinical characteristics of the HIV-1 infected patients and healthy population included in the study.
CTLA-4 (+49A/G) polymorphism and HIV-1 infection
The distribution and comparison of genotype and allele frequencies of CTLA-4 +49 G/A polymorphism in cases and controls are shown in Table 2. The observed genotype frequencies were in concordance with the Hardy-Weinberg equilibrium in both cases(P=0.970) and controls (P=0.619). Individuals who carried AA homozygosity were 57 (60.00%), AG heterozygous were 30 (31.58%) and GG homozygous were 8 (8.42%), suggesting that AA and AG genotypes were more likely to be the chief genotypes for Han Chinese than those who carried the GG genotypes at the CTLA-4 polymorphism, +49 A>G. Genotype frequencies in HIV-1 infected cases and healthy controls for AA were 53.33% and 66.00%; for AG were 37.78% and 26.00%; for GG were 8.89% and 8.00%, for the combined genotype AG+GG were 46.67% and 34.00%, respectively. The OR for the AG genotype was 1.80 (95% CI=0.74-4.39); for the GG genotype was 1.32 (95% CI=0.36-4.83); for the combined genotype AG and GG was 1.70 (95% CI=0.74-3.89), no statistically significant differences were noted (P>0.05). The frequency of the G allele did not differ between cases (G allele present in 27.78% whereas A allele present in 72.22%) and controls (G allele present in 21.00% whereas A allele present in 79.00%). No significant differences in the frequencies of alleles were found between groups (P=0.276). The OR for the G allele was 1.45 (95% CI=0.74-2.82), implicating that G allele may not be one of the major candidate genes for HIV-1 infection.
| CTLA-4 +49 A → G SNP | HIV-1 patients (No. (%)) |
Healthy controls (No. (%)) |
χ2 (P value*) | OR (95% CI) |
|---|---|---|---|---|
| A | 65 (72.22) | 79 (79.00) | 1 | |
| G | 25 (27.78) | 21 (21.00) | χ2=1.19 (P=0.276) | 1.45 (0.74?2.82) |
| AA | 24 (53.33%) | 33 (66.00%) | 1 | |
| AG | 17 (37.78%) | 13 (26.00%) | χ2=1.67 (P=0.196) | 1.80 (0.74?4.39) |
| GG | 4 (8.89%) | 4 (8.00%) | χ2=0.178 (P=0.673) | 1.38 (0.31?6.05) |
| AG and GG | 21 (46.67%) | 17 (34.00%) | χ2=1.58 (P=0.208) | 1.70 (0.74?3.89) |
| PHWE=0.970 | PHWE=0.619 |
*Two-sided Fisher’s exact or χ2.P<0.05 was required for statistical significance and is presented in bold.
Table 2. Allele and genotype distribution of the CTLA-4 A+49G SNP in 45 HIV-1-Infected Cases vs. 50 Healthy controls of Han Chinese.
Initial plasma viral load, CD+4 T-cell counts, CD +4/CD+8 ratios according to genotypes
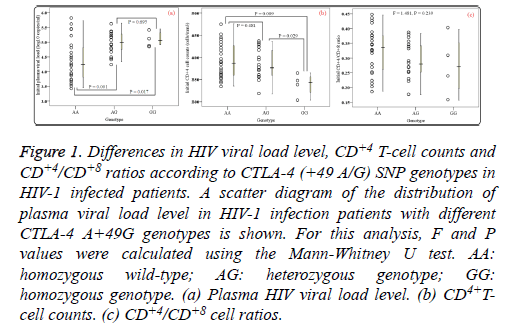
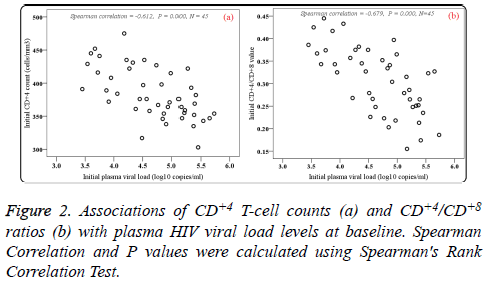
Of the patients with HIV-1 infection, 24 (53.33%) were wildtype (A/A), 17 (37.78%) were heterozygous variants (A/G), and 4 (8.89%) were homozygous variants (G/G). PHVL, CD+4 T-cell counts and CD+4/CD+8 ratios at baseline were observed. As shown in Figures 1a and 1b, there were statistical significance in mean PHVL and CD+4 T-cell counts among the three genotypes (P<0.05). Significant differences were observed between AA and GG genotypes (P<0.05), however, there was no difference in PHVL between AG and GG genotypes (P=0.695). Similar results were found in CD+4 Tcell counts between AA and AG genotypes. As to CD+4/CD+8 ratios, our study showed no difference in either genotype (F=1.481, P=0.239) (Figure 1c). Moreover, the relationships between initial CD+4 T-cell counts and CD+4/CD+8 ratios values and initial PHVL (log 10 copies/ml) were shown as a scatter dot plot graph in the 45 samples (Figure 2). With the initial PHVL increasing, both initial CD+4 T-cell counts and CD+4/CD+8 ratios were decreasing, indicating they had a negative correlation (P<0.001).
Figure 1: Differences in HIV viral load level, CD+4 T-cell counts and CD+4/CD+8 ratios according to CTLA-4 (+49 A/G) SNP genotypes in HIV-1 infected patients. A scatter diagram of the distribution of plasma viral load level in HIV-1 infection patients with different CTLA-4 A+49G genotypes is shown. For this analysis, F and P values were calculated using the Mann-Whitney U test. AA: homozygous wild-type; AG: heterozygous genotype; GG: homozygous genotype. (a) Plasma HIV viral load level. (b) CD4+Tcell counts. (c) CD+4/CD+8 cell ratios.
Comparison of the plasma EFV concentrations in the three genotype groups
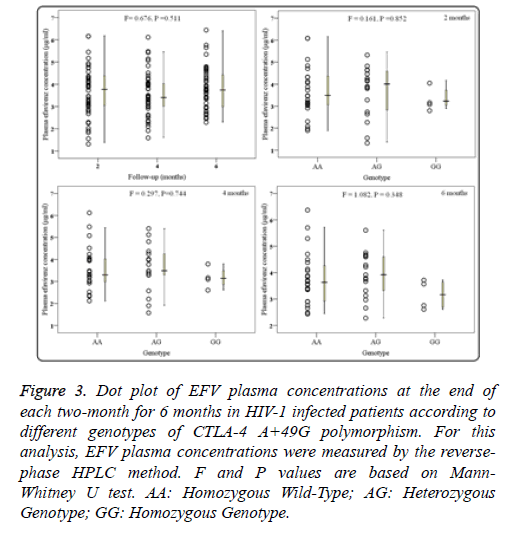
HIV-1 infection patients receiving EFV/AZT/3TC treatment were followed up for 6 months, and data of plasma EFV concentrations were collected at the end of every two months (2, 4, and 6 months). As shown in Figure 3a, significant differences in overall plasma concentrations of EFV were not observed among every two months (F=0.676, P=0.511). Moreover, groups of different +49A/G genotypes showed no apparent change on the plasma EFV concentrations at 2 months (F=0.161, P=0.852; Figure 3b). Similarly, there was no apparent difference among three genotypes at 4 months and 6 months (P>0.1; Figures 3c and 3d).
Figure 3: Dot plot of EFV plasma concentrations at the end of each two-month for 6 months in HIV-1 infected patients according to different genotypes of CTLA-4 A+49G polymorphism. For this analysis, EFV plasma concentrations were measured by the reversephase HPLC method. F and P values are based on Mann- Whitney U test. AA: Homozygous Wild-Type; AG: Heterozygous Genotype; GG: Homozygous Genotype.
Association between CTLA-4 (+49A/G) polymorphism and clinical response
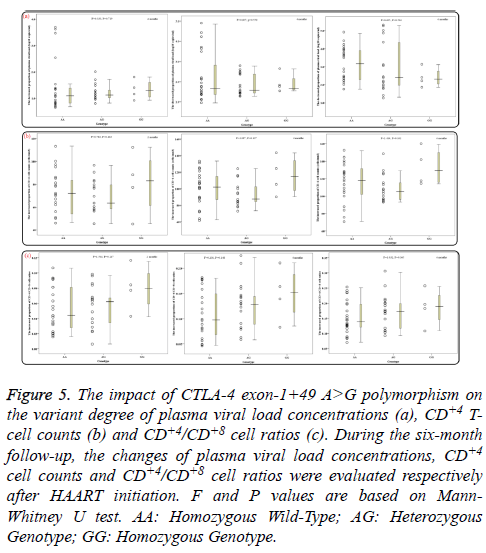
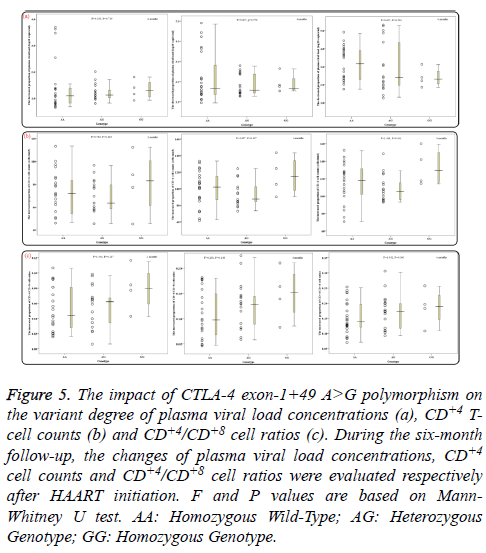
During the six-month follow-up, there were statistically significant differences in mean levels of overall PHVL, CD+4 T-cell counts and CD+4/CD+8 ratios between every two months (all P<0.02). With increasing HAART time, the PHVL levels (log 10 copies/ml) were decreased from 4.67 ± 0.63 to 3.35 ± 1.08, 2.66 ± 1.21 and 1.69 ± 1.38 respectively, the differences were statistically significant (P<0.001; Figure 4a), whereas the mean CD+4 T-cell counts and the CD+4/CD+8 ratios were increased significantly (P<0.001; Figures 4b and 4c). Additionally, groups of AA, AG and GG genotypes showed no notable difference in the decreased proportion of PHVL among every 2 months (for 2months, P=0.719; for 4 months, P=0.550; for 6 months, P=0.504; Figure 5a). Similar results, according to different genotypes, were found in the increment proportion of CD+4 T-cell counts and CD+4/CD+8 ratios among every 2 months (P >0.1; Figures 5b and 5c).
Figure 5: The impact of CTLA-4 exon-1+49 A>G polymorphism on the variant degree of plasma viral load concentrations (a), CD+4 Tcell counts (b) and CD+4/CD+8 cell ratios (c). During the six-month follow-up, the changes of plasma viral load concentrations, CD+4 cell counts and CD+4/CD+8 cell ratios were evaluated respectively after HAART initiation. F and P values are based on Mann- Whitney U test. AA: Homozygous Wild-Type; AG: Heterozygous Genotype; GG: Homozygous Genotype.
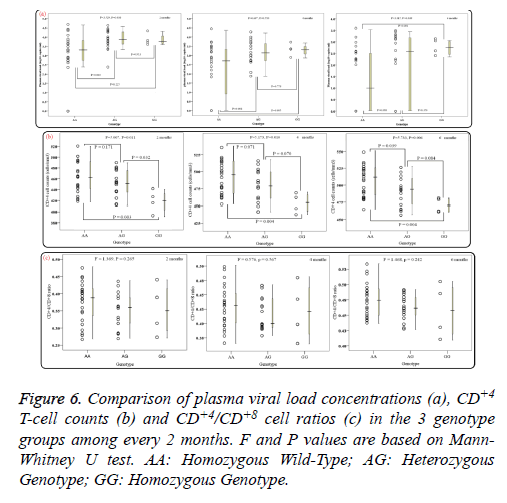
Of note, in 12 patients of the AA genotype no PHVL could be detected, including 3 patients were for the 2 months, 3 for the 4 months and 6 for the 6 month. In addition, 4 patients for the 6 months of the AG genotype were undetectable as well. When data of PHVL from above total 25 patients was excluded for further analysis, interestingly, there was apparent difference in mean PHVL between AA genotype and AG genotype for the 2 and 4 months (P<0.05; Figure 6a), moreover, significant difference was also observed between AA genotype and GG genotype for the 4 and 6 months (P<0.05). As for the CD+4 Tcell counts, there were significant differences between AA genotype and GG genotype among every 2 months (P<0.05; Figure 6b). However, for the CD+4/CD+8 ratios among every 2 months, no statistical significance was observed (P>0.05; Figure 6c), according to different genotypes.
Discussion
HIV-1 infection causes a progressive impairment of the immune system, characterized by a massive CD+4 T-cell depletion, and sustained immune activation and inflammation. CTLA-4 is a surface marker expressed on regulatory T cells (Tregs) and activated T lymphocytes [32]. Upregulation of CTLA-4 appears closely linked to HIV replication and progressive disease [33]. It has been shown that SNPs in CTLA-4 may have functional significance. The +49 A allele, which causes an amino acid exchange (from alanine to threonine) in the peptide leader sequence, results in higher mRNA efficiency and more CTLA-4 production than the +49 G allele [34]. In animal studies, the GG genotype of CTLA-4 (+49) A/G polymorphism was associated with lower inhibitory function, and the G allele was associated with lower CTLA-4 expression [35]. Genes encoding proteins involved in T-cell activation and suppression have been considered as candidate gene for many autoimmune and neoplastic diseases [13,15], maybe, also for HIV-1 infection. In the present study, we firstly attempt to examine the association of CTLA-4 gene polymorphism A/G in exon 1 (+49) with HIV-1 infection susceptibility. Unfortunately, we did not observe significant differences in allele and genotype frequencies of the CTLA-4+49A/G polymorphism between HIV-1 infected patients and controls. It should be noted, however, that the number of HIV-1 infected patients in this study was small (n=45).
activation that promotes viral replication and plays a central role in HIV/AIDS progression. Chronic immune activation is overwhelmingly detrimental, it results in the generation of activated T cells that are targets for the HIV, thus further driving viral replication and CD+4 T-cell depletion [36]. In theory, an alanine to threonine change of the +49 A/G polymorphism in the leader sequence, which is later processed in the endoplasmic reticulum, may result in reduced expression of membrane CTLA-4 protein and enhance T cell proliferative response [37,38]. An unexpected finding in current study, which is difficult to explain, is that CTLA-4 +49 A/G polymorphism was associated with PHVL level. Patients of AA genotype were associated with lower PHVL and higher CD+4 T-cell compared to GG genotype patients. It is possible that polymorphic modifications of CTLA-4 gene resulting in a T-cell membrane receptor less effective in suppressor function would start immune activation, which is associated with more rapid clinical progression and CD+4 T-cell declines [39]. Since the +49 G allele has been associated with lower protein expression than the +49 A allele, and therefore reduced the control of activation and pronounced proliferation of T cells, producing enhanced immune response could lead to the altered T cell regulation [12,40], which might contribute to obviously enhanced HIV virus load. Consequently, in addition to the implication that the genetically predisposed susceptibility involving CTLA-4 polymorphism affects the outcome of HIV-1 infection, the data of CTLA-4 gene polymorphism in HIV-1 infection may provide an immunologic rationale for immunotherapeutic designs that combine antiviral therapy with B7-CD28/ CTLA-4 blockade based on genetic background of the HIV-1-infected patient.
Immune activation and erosion of the naive T cell pool is strongly associated with HIV-1 infection. The introduction of HAART has led to very important declines in both mortality and morbidity due to HIV infection. HAART can employ several different drugs. Actually, the use of several drugs is essential to help prevent the rise of resistant HIV strains [41,42]. EFV, which is a first generation non-nucleoside reverse transcriptase inhibitor of HIV-1 and is one of the preferred components of the first line treatment regimen of HIV infection worldwide, has the potential of preserving high adherence rates necessary to achieve sustained virologic suppression [43]. Plasma concentrations of EFV exhibit large inter-patient variability. Ethnicity, sex, body weight, underlying disease condition, and genetic factors are all associated with EFV plasma concentration. With total EFV plasma concentrations related to viral response and drug toxicity, investigation of factors having an impact on the intracellular and plasma concentrations of EFV is warranted. Association between polymorphisms in the CYP2B6 gene and EFV plasma concentrations was well documented [31,44]. However, this association is not sufficient to explain all the pharmacokinetic variability seen in patients. To date, little is known about the association between CTLA-4 exon 1 polymorphism (+49A/G) and plasma EFV concentration. In present study, although our findings suggested that SNP in CTLA-4 exon 1 (+49A>G) was not associated with plasma EFV concentration, interestingly, the plasma EFV level of GG genotype was the lowest during 6 months of HAART, suggesting therapeutic dose of EFV may need to be raised for the GG genotype patients. The information given by this single SNP analysis may help to easily identify HIV infection patients who have a risk for treatment failure. We consider that the association between CTLA-4 (+49) A/G polymorphisms and low EFV concentration urgently needs further studies.
Regarding clinical response, high levels of Tregs have been linked with suppression of HIV-specific CD+4 and CD+8 T-cell activities. The CD+4 T-cell counts is an explicit biomarker that provides assessment of immune system status of HIV-infected patients, while plasma HIV virus load reflects activity of HIV virus [45,46]. As expected, current study showed that EFVcontaining HAART led to improvement in CD+4 T-cell counts, increase in CD+4/CD+8 ratios and PHVL decline when baseline levels were compared to levels at 6 months post- HAART initiation, reflecting inhibition of HIV replication and restoration of CD+4 and CD+8 T-cell subsets and function. We have also observed a strong and inverse correlation between PHVL and CD+4 T-cell counts in HIV-1 infection patients, suggesting that factors independent of viral RNA load are driving CD+4 T-cell activation. In HIV-1 infection, low viral RNA loads usually predict good CD+4 T-cell counts [47]. Present data supports that activation or exhaustion of T cells is associated with the ability to produce these specific cytokines and that the impairment in T cell function is reversible.
Moreover, we report here there was no significant difference in changes of HIV virus load, CD+4 T-cell counts and CD+4/CD +8 ratios in either genotype for 2, 4 and 6 months. Intriguingly, 16 patients (including 75% of AA genotype and 25% of AG genotype) presented HIV-1 PHVL below the minimum detectable level threshold, and if above patients were excluded for further analysis, there was significant difference in mean PHVL between AA and GG genotypes for the 4 and 6 months. Similar results were found in CD+4 T-cell counts among every 2 months, suggesting the possibility that the observed relationship may result in part from direct effects on HIV function. Because +49 G allele has been associated with higher PHVL than the +49 A allele, which has been found in this study, and HIV-1 establishes a systemic infection with high PHVL that exhibits pathogenic properties by depleting human CD+4 T-cells [39,46]. Taken together, it is worth noting that there was non-association between genetic variation in CTLA-4 exon 1 (+49 A>G) and Treg response to EFV-based HAART regimens.
In conclusion, we found that the CTLA-4 (+49) A/G polymorphisms had no association with the susceptibility of HIV-1 infection, but the CTLA-4 +49G allele was significantly associated with PHVL level in HIV-1 infection. However, the small sample size in our study may lead to a relatively lower statistical power, so further studies should include larger sample sizes for replication and the roles of haplotypes of the CTLA-4 (+49) gene in the accumulation of HIV virus load required to be clarified in the future researches.
Conflicts of Interests
None
References
- World Health Organization. HIV/AIDS. 20th International AIDS Conference: Stepping up the pace, 2014.
- Ncaids N, China CDC. Update on the AIDS/STD epidemic in China and main response in control and prevention in February, 2014. Chin J AIDS STD 2014; 20: 227.
- Demers KR, Reuter MA, Betts MR. CD8 (+) T-cell effector function and transcriptional regulation during HIV pathogenesis. Immunol Rev 2013; 254: 190-206.
- Ghezzi S, Galli L, Kajaste-Rudnitski A, Turrini F, Marelli S. Identification of TRIM22 single nucleotide polymorphisms associated with loss of inhibition of HIV-1 transcription and advanced HIV-1 disease. AIDS 2013; 27: 2335-2344.
- Larsson M, Shankar EM, Che KF, Saeidi A, Ellegard R. Molecular signatures of T-cell inhibition in HIV-1 infection. Retrovirology 2013; 10: 31.
- Vaccari M, Boasso A, Fenizia C. Fatal pancreatitis in simian immunodeficiency virus SIV(mac251)-infected macaques treated with 2,3-dideoxyinosine and stavudine following cytotoxic-T-lymphocyte-associated antigen 4 and indoleamine 2,3-dioxygenase blockade. J Virol Jan 2012; 86: 108-113.
- Che KF, Shankar EM, Muthu S. p38 Mitogen-activated protein kinase/signal transducer and activator of transcription-3 pathway signalling regulates expression of inhibitory molecules in T cells activated by HIV-1-exposed dendritic cells. Mol Med 2012; 18: 1169-1182.
- Tai X, Van Laethem F, Pobezinsky L, Guinter T, Sharrow SO. Basis of CTLA-4 function in regulatory and conventional CD4 (+) T cells. Blood 2012; 119: 5155-5163.
- Stojanovic A, Fiegler N, Brunner-Weinzierl M, Cerwenka A. CTLA-4 is expressed by activated mouse NK cells and inhibits NK Cell IFN-γ production in response to mature dendritic cells. J Immunol 2014; 192: 4184-4191.
- Frydecka D, Beszlej A, Szewczuk-Boguslawska M. The role of genetic variations of immune system regulatory molecules CD28 and CTLA-4 in schizophrenia. Psych Res 2013; 208: 197-198.
- Liu P, Xu L, Sun Y, Wang ZP. The association between cytotoxic T lymphocyte-associated antigen-4 and cervical cancer. Tumor Biol 2014; 35: 2893-2903.
- Wang L, Su G, Zhao X, Cai Y, Cai X. Association between the cytotoxic T-lymphocyte antigen 4 +49A/G polymorphism and bladder cancer risk. Tumour Biol 2014; 35: 1139-1142.
- Danilovic DL, Mendes-Correa MC, Lima EU, Zambrini H, K Barros R. Correlations of CTLA-4 gene polymorphisms and hepatitis C chronic infection. Liver Int 2012; 32: 803-808.
- Vaidya B, Imrie H, Perros P. Cytotoxic T lymphocyte antigen-4 (CTLA-4) gene polymorphism confers susceptibility to thyroid associated orbitopathy. Lancet1999; 354: 743-744.
- Kavvoura FK, Akamizu T, Awata T. Cytotoxic T-lymphocyte associated antigen 4 gene polymorphisms and autoimmune thyroid disease: A meta-analysis. Journal of Clin Endocrinol Metabol 2007; 92: 3162-3170.
- Tang MJ, Zhou ZB. Association of the CTLA-4 +49A/G polymorphism with rheumatoid arthritis in Chinese Han population. Mol Biol Rep 2013; 40: 2627-2631.
- Haque R, Lei F, Xiong X, Bian Y, Zhao B. Programming of regulatory T cells from pluripotent stem cells and prevention of autoimmunity. J Immunol 2012; 189: 1228-1236.
- Yang KD, Yang MY, Li CC, Lin SF, Chong MC, Wang CL, Chen RF, Lin TY. Altered cellular but not humoral reactions in children with complicated enterovirus 71 infections in Taiwan. J Infect dis 2001; 183: 850-856.
- Tomoyose T, Komiya I, Takara M. Cytotoxic T-lymphocyte antigen-4 gene polymorphisms and human T-cell lymphotrophic virus-1 infection: Their associations with Hashimotos thyroiditis in Japanese patients. Thyroid 2002; 12: 673-677.
- Jiang ZJ, Feng XN, Zhang W. Recipient cytotoxic T lymphocyte antigen-4+49 G/G genotype is associated with reduced incidence of hepatitis B virus recurrence after liver transplantation among Chinese patients. Liver Int 2007; 27: 1202-1208.
- Zhang GY, Han QY, Duan SQ. PDCD1 polymorphism amplifies the predisposing effect conferred by CTLA4 polymorphism in chronic hepatitis B virus infection. Human Immunology 2012; 73: 421-425.
- Toth I, Le AQ, Hartjen P. Decreased frequency of CD73 (+) CD8 (+) T cells of HIV-infected patients correlates with immune activation and T cell exhaustion. J Leuk Biol 2013; 94: 551-561.
- Vollbrecht T, Stirner R, Tufman A, Roider J, Huber RM. Chronic progressive HIV-1 infection is associated with elevated levels of myeloid-derived suppressor cells. AIDS 2012; 26: F31-37.
- Heath SL, Sabbaj S, Bansal A, Kilby JM, Goepfert PA. CD8 T-cell proliferative capacity is compromised in primary HIV-1 infection. J Acquir Immune Defic Syndr 2011; 56: 213-221.
- Whittall T, Peters B, Rahman D, Kingsley CI, Vaughan R, Lehner T. Immunogenic and tolerogenic signatures in human immunodeficiency virus (HIV)-infected controllers compared with progressors and a conversion strategy of virus control. Clin Exp Immunol 2011; 166: 208-217.
- Zur Wiesch JS, Thomssen A, Hartjen P. Comprehensive analysis of frequency and phenotype of t regulatory cells in hiv infection: CD39 expression of FoxP3 (+) T regulatory cells correlates with progressive disease. J Virol 2011; 85: 1287-1297.
- Pollard RB, Rockstroh JK, Pantaleo G. Safety and efficacy of the peptide-based therapeutic vaccine for HIV-1, Vacc-4x: a phase 2 randomised, double-blind, placebo-controlled trial. Lancet Infect Dis 2014; 14: 367.
- Campbell TB, Smeaton LM, Kumarasamy N. Efficacy and safety of three antiretroviral regimens for initial treatment of HIV-1: A randomized clinical trial in diverse multinational settings. Plos Medicine 2012; 9.
- Avery LB, Sacktor N, McArthur JC, Hendrix CW. Protein-free Efavirenz concentrations in cerebrospinal fluid and blood plasma are equivalent: applying the law of mass action to predict protein-free drug concentration.Antimicrob Agent Chemother 2013; 57: 4095.
- Rotger M, Colombo S, Furrer H. Influence of CYP2B6 polymorphism on plasma and intracellular concentrations and toxicity of efavirenz and nevirapine in HIV-infected patients. Pharmacog Genom 2005; 15: 1-5.
- Gandhi M, Greenblatt RM, Bacchetti P. A single-nucleotide polymorphism in CYP2B6 leads to>3-fold increases in efavirenz concentrations in plasma and hair among HIV-infected women. J Infect Dis 2012; 206: 1453-1461.
- Leligdowicz A, Feldmann J, Jaye A, Cotten M, Dong T. Direct relationship between virus load and systemic immune activation in HIV-2 infection. J Infect Dis 2010; 201: 114-122.
- Singh M, Singh P, Vaira D, Amand M, Rahmouni S. Minocycline attenuates HIV-1 infection and suppresses chronic immune activation in humanized NOD/LtsZ-scidIL-2Rγ (null) mice. Immunol 2014; 142: 562-572.
- Wang W, Wang J, Song H, Liu J, Song B. Cytotoxic T-lymphocyte antigen-4 +49G/A polymorphism is associated with increased risk of osteosarcoma. Genet Test Mol Biomarkers 2011; 15: 503-506.
- Kuo HC, Liang CD, Yu HR, Wang CL, Lin IC. CTLA-4, position 49 A/G polymorphism associated with coronary artery lesions in Kawasaki disease. J Clin Immunol 2011; 31: 240-244.
- Moir S, Chun TW, Fauci AS. Pathogenic mechanisms of HIV disease. Annu Rev Pathol 2011; 6: 223-248.
- Takara M, Komiya I, Kinjo Y, Tomoyose T, Yamashiro S. Association of CTLA-4 gene A/G polymorphism in Japanese type 1 diabetic patients with younger age of onset and autoimmune thyroid disease. Diabetes Care 2000; 23: 975-978.
- Agarwal K, Jones DE, Daly AK, James OF, Vaidya B. CTLA-4 gene polymorphism confers susceptibility to primary biliary cirrhosis. J Hepatol 2000; 32: 538-541.
- Groves KC, Bibby DF, Clark DA, Isaksen A, Deayton JR. Disease progression in HIV-1-infected viremic controllers. J Acquir Immune Defic Syndr 2012; 61: 407-416.
- Yang M, Sun T, Zhou Y, Wang L, Liu L. The functional cytotoxic T lymphocyte-associated Protein 4 49G-to-A genetic variant and risk of pancreatic cancer. Cancer 2012; 118: 4681-4686.
- Cohen C, Simonsen L, Sample J. Influenza-related mortality among adults aged 25-54 years with AIDS in South Africa and the United States of America. Clin Infect Dis 2012; 55: 996-1003.
- Le Douce V, Janossy A, Hallay H, Ali S, Riclet R. Achieving a cure for HIV infection: do we have reasons to be optimistic? J Antimicrob Chemother 2012; 67: 1063-1074.
- Mackie NE, Dunn DT, Dolling D. The impact of HIV-1 reverse transcriptase polymorphisms on responses to first-line non-nucleoside reverse transcriptase inhibitor-based therapy in HIV-1-infected adults. Aids 2013; 27: 2245-2253.
- Manosuthi W, Sukasem C, Thongyen S, Nilkamhang S, Manosuthi S, Sungkanuparph S. CYP2B6 18492T->C polymorphism compromises efavirenz concentration in co-infected HIV and tuberculosis patients carrying CYP2B6 haplotype1/1. Antimicrob Agents Chemother 2014; 58: 2268-2273.
- Mendez-Lagares G, Pozo-Balado MM, Genebat M, Garcia Perganeda A, Leal M, Pacheco YM. Severe immune dysregulation affects CD4 (+) CD25 (hi) FoxP3 (+) regulatory T cells in HIV-infected patients with low-level CD4 T-cell repopulation despite suppressive highly active antiretroviral therapy. J Infect Dis 2012; 205: 1501-1509.
- Manzke N, Akhmetzyanova I, Hasenkrug KJ, Trilling M, Zelinskyy G, Dittmer U. CD4+ T cells develop antiretroviral cytotoxic activity in the absence of regulatory T cells and CD8+ T cells. J Virol 2013; 87: 6306-6313.
- Frater J, Ewings F, Hurst J, Brown H, Robinson N. HIV-1-specific CD4 (+) responses in primary HIV-1 infection predict disease progression. AIDS 2014; 28: 699-708.