ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2016) Volume 27, Issue 4
Analyzed the genotypic and phenotypic antibiotic resistance patterns of Klebsiella pneumoniae isolated from clinical samples in Iran
1Cellular and Molecular Research Center, Sabzevar University of Medical Sciences, Sabzevar, Iran
2Department of Microbiology, Faculty of Basic Sciences, Shahrekord Branch, Islamic Azad University, Shahrekord, Iran
- *Corresponding Author:
- Elahe Tajbakhsh
Department of Microbiology
Faculty of Basic Sciences
Shahrekord Branch
Islamic Azad University
Iran
Accepted date: March 21, 2016
Klebsiella pneumoniae is one of the most important opportunistic enteric bacteria and is a major cause of pneumonia and urinary tract infection. Serotype capsules of K1 and K2 can cause intense diseases. Acquisition of plasmid that codes the production of ESBLs confers on K. pneumoniae resistance to number of broad spectrum antibiotics posing a global public health problem. Integron is one of the important factors of multi resistance in gram negative microorganism’s especially intestinal bacteria. The magA gene rmpA gene was studied in 90 isolates of K. pneumoniae from different clinical cases in Shahrekord city, Iran. The frequency of resistance genes qnr, sul 1, tetB, tetA and aac (3) IIa at the presence of specific primers were examined and all resistant isolates were tested for detection of sul1, sul2, sul 3 and int1 genes using special primers. Of the 90 isolates, 13 had serotype K1A with redundancy of 14.44% and 15 cases had serotype K2A with the redundancy of 16.66%. rmpA gene was observed in 10 isolates the redundancy of 11.11%. In this study 33 isolates resistance to cotrimoxazole, aren’t finding sul1 gene in 15 isolated cases, sul2 gene in 20 isolated cases, sul3 gene in 2 isolated cases, respectively. Also there were 27 demonstrating int1 genes for Cotrimoxazol. The study has revealed that serotype K1 is one of the most important serotypes of K. pneumonia. Also there seems to be a strong relationship between presence of Integron and increased resistance to different antibiotics.
Keywords
Antibiotic resistance genes, Capsular antigen, Integron, Klebsiella pneumoniae, rmpA gene.
Introduction
Klebsiella pneumoniae is a gram-negative, aerobic, non motile bacillus and is a common cause of a wide range of infections in humans and animals [1,2] and one of the most common enteric bacteria responsible for up to 10% of all nosocomial infections and also involved in pneumonia and urinary tract infections causing severe morbidity and mortality [1,3]. Recently, a highly invasive K. pneumoniae causing primary liver abscesses in humans has also been reported [4-6]. These invasive, abscess-forming strains of K. pneumoniae are associated with the so-called hypermucoviscosity (HMV) phenotype, a bacterial colony trait identified by a positive string test [7-9]. The HMV phenotype is seen in K. pneumoniae expressing either the capsular serotypes K1 or K2. K1 serotypes of K. pneumoniae have 2 potentially important genes, rmpA, a transcriptional activator of colanic acid biosynthesis, [10] and magA, which encodes a 43-kD outer membrane protein [7]. K2 serotypes of K. pneumoniae also have rmpA but do not have magA. Serotype capsules of K1 and K2 can cause intense diseases and based on studies of these serotypes, it has been revealed that magA gene, related to Hypermocoviscosities and rmpA gene, in charge of positive synthesis of outside-capsule polysaccharide, are both useful tools in knowing such serotypes. Most K. pneumoniae isolates have a chromosomally encoded SHV-1 β-lactamase [11]. Since 1983, plasmid-encoded extended-spectrum β-lactamases (ESBLs) derived from the TEM and SHV families have been extensively reported in Enterobacteriaceae, especially in Klebsiella spp. [12,13].
Emergence and spread of multidrug resistant K. pneumoniae, specifically the ESBL producing strains, is often responsible for the failure of antibiotic treatment in hospital settings [14]. In many countries, however, the presence of resistance to Trimethoprim-sulfamethoxazole can lead to treatment failure in cases of UTIs [15]. Sulfonamide resistance in gram-negative bacilli generally arises from the acquisition of dihydropteroate synthase (DHPS) genes in integrons that are not inhibited by the drug [16]. Currently, three different types of DHPS genes (sul1, sul2, and sul3) are known [15]. The sul1 gene is found linked to other resistance genes in class 1 integrons and on large conjugative plasmids [17], while sul2 is usually located on small nonconjugative plasmids [18], large transmissible multi-resistance plasmids [15], or through insertion element common region (ISCR2) element [19]. Although rare, sul3, a plasmid-borne sulfonamide resistance gene, is also present [17].
Recent studies have shown that mobile and mobilizable DNA elements, such as integrons, play an important role in the development and dissemination of antibiotic resistance [20-22]. Integrons are defined as site-specific recombination systems that are capable of integrating and expressing open reading frames contained in modular structures called mobile gene cassettes [23]. Different classes of integrons are characterized by sequence differences in the intI gene encoding an integrase. Class 1 integrons possess two conserved segments (CSs), the 5'-CS and the 3'-CS, separated by a variable region, which includes integrated antibiotic resistance gene cassettes of different lengths, arrangements, and sequences [23]. In the clinical environment, three main groups or classes of integrons associated with antibiotic resistance have been described. Class 1 integrons are most frequently found in clinical isolates of Gram-negative bacteria [22]. Although several literatures studied sul and/or dfr genes in relation to class 1 integron in E. coli [15,24-26], there are limited reports investigating the antigenic capsules of K. pneumoniae, the phenotypic genotypic antibiotic resistance patterns in K. pneumoniae and sul genes in relation to class 1 integrons and sul genes in Klebsiella in Iran. Therefore, in this present study, we investigated the genotypic and phenotypic antibiotic resistance patterns of strains of K. pneumoniae isolated from clinical samples in Iran
Materials and Methods
Bacterial strains and identification
We examined 90 K. pneumoniae clinical isolates from hospitals of Shahrekord, Iran. Clinical isolates were mostly from urine, blood culture, eye secretion, trachea and wound. Prior to molecular-serotyping, all clinical isolates were biochemically identified by conventional bacteriology tests as detailed previously [27].
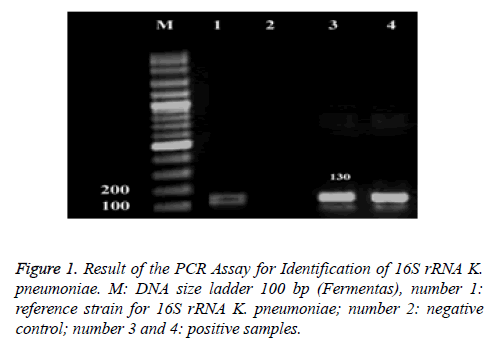
The PCR method was used to detect the 16S–23S internal transcribed spacer unit of K. pneumoniae subsp. pneumoniae, facilitating identification of this subspecies, as described previously [28]: F: ATTTGAAGAGGTTGCAAACGAT and R: TTCACT CTGAAGTTTTCTTGTTTC (amplicon size: 130 bp). Cycling conditions were as follows: Initial denaturation at 94°C for 5 min; 35 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 1 min followed by a final extension at 72°C for 7 min. K. pneumoniae ATCC13883 was used as positive control.
Antimicrobial susceptibility testing
The antibiotic susceptibility was determined by disk diffusion method on Mueller-Hinton agar plates (Merck, Darmstadt, Germany) as recommended by the Clinical Laboratory Standards Institute (CLSI) [29]. The disks containing the following antibiotics were used (Padtan-Teb, Iran): amoxiciline (10 μg), amikacin (30 μg), kanamycin (30 μg), tetracycline (30 μg), nalidixic acid (30 μg), co-trimoxazole (25 μg), ciprofloxacin (5 μg), cephalothin (30 μg), norfloxacin (10 μg), ceftriaxone (30 μg), nitrofurantoin (10 μg), imipenem (10 μg), cefepime (30 μg), and gentamicin (10 μg). E. coli ATCC 25922 was used as quality control for antimicrobial susceptibility test.
Polymerase chain reaction assay
The DNA template was extracted using phenol and chloroform method. The total DNA was measured at 260 nm optical density according to the method described by Sambrook and Russell [30]. The reverse and forward primers, size of product and PCR programs (temperature and volume) as previously published used for the detection of capsular K1 and K2 serotypes in K. pneumoniae in this study are presented in Table 1 [28]. In addition, The primers, size of product and PCR conditions as previously published used for the detection of resistant genes and sul genes of K. pneumoniae are presented in Table 2 and Table 3, respectively [15,31-34]. Reference strains of K. pneumoniae AY762939 and K. pneumoniae D21242 were used as positive controls for PCR reactions of K1 and K2 serotypes respectively.
| Gene | Primer name | Primer Sequence (5'-3') | Size of product (bp) | PCR program | PCR volume (50 µl) |
Reference |
|---|---|---|---|---|---|---|
| K1A | aac(3)-IV | (F) GGTGCTCTTTACATCATTGC (R) GCAATGGCCATTTGCGTTAG |
1283 | 1 cycle: 95°c ------------ 10 min. 34 cycle: 95°c ------------ 30 s 58°c ------------ 60 s 72°c ------------ 90 s 1 cycle: 72°c ------------ 5 min |
5 µl PCR buffer 10X 2.5 mm Mgcl2 200 µM dNTP (Fermentas) 0.5 µm of each primers F & R 2 U Taq DNA polymerase (Fermentas) 3 µl DNA template |
[28] |
| K2A | sul1 | (F) GACCCGATATTCATACTTGACAGAG (R)CCTGAAGTAAAATCGTAAATAGATGGC |
641 | |||
| rmpA | blaSHV | (F) ACTGGGCTACCTCTGCTTCA (R) CTTGCATGAGCCATCTTTCA |
536 | |||
| F- Forward; R- Reverse | ||||||
Table 1: Primers used for genes in K. pneumoniae.
| Antibiotic | Resistant gene | Sequence | Size (bp) | Anealing | PCR program | References |
|---|---|---|---|---|---|---|
| Tetracycline | tetA | GTGAAACCCAACATACCCC GAAGGCAAGCAGGATGTAG |
888 | 55 | 1 cycle: 94°c ------------ 5 min cycle: 30 49°c ------------ 15 s 55°c ------------ 60 s 72°c ------------ 60 s 1 cycle: 72°c ------------ 5 min |
[31] |
| Tetracycline | tetB | CCTTATCATGCCAGTCTTGC ACTGCCGTTTTTTCGCC |
774 | 55 | 1 cycle: 95°c ------------ 5 min. 30 cycle: 95°c ------------ 30 s 55°c ------------ 60 s 72°c ------------ 60 s 1 cycle: 72°c ------------ 5 min |
|
| Fluoroquinolone | qnr | ATTTCTCACGCCAGGATTTG GATCGGCAAAGGTTAGGTCA |
516 | 55 | 1 cycle: 95°c ------------ 5 min. 30 cycle: 95°c ------------ 30 s 55°c ------------ 60 s 72°c ------------ 60 s 1 cycle: 72°c ------------ 5 min |
[32] |
| Gentamicin | aac(3)IIa | CGGAAGGCAATAACGGAG TCGAACAGGTAGCACTGAG |
740 | 55 | 1 cycle: 94°c ------------ 5 min cycle: 30 49°c ------------ 15 s 55°c ------------ 60 s 72°c ------------ 60 s 1 cycle: 72°c ------------ 5 min |
[31] |
| Sulfonamide | Sul1 | CGGCGTGGGCTACCTGAACG GCCGATCGCGTGAAGTTCCG | 433 | 65 | 1 cycle: 94°c ------------ 5 min. 34 cycle: 94°c ------------ 60 s 65°c ------------ 60 s 72°c ------------ 90 s 1 cycle: 72°c ------------ 8 min |
[33] |
Table 2: Primers and PCR conditions of resistant genes of K. pneumoniae.
| Gene | Sequence (5’–3’) | Annealing temp (°C) | Size of product (bp) | Reference |
|---|---|---|---|---|
| Sul 1 | F: CGGCGTGGGCTACCTGAACG | 65 | 433 | [15] |
| R: GCCGATCGCGTGAAGTTCCG | ||||
| Sul 2 | F: GCGCTCAAGGCAGATGGCATT | 65 | 293 | [15] |
| R: GCGTTTGATACCGGCACCCGT | ||||
| Sul 3 | F: GCCTATGCATCTACACAATC | 65 | 750 | [34] |
| R: TGAGAAATGGACAATGTCCG | ||||
| Int1 | F: CAGTGGACATAAGCCTGTTC | 53 | 160 | [34] |
| R: CCCGAGGCATAGACTGTA |
Table 3: Primers used for sul genes.
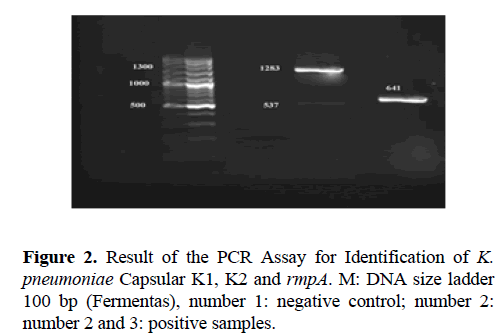
The 2% agarose gel in TBE buffer was used for PCR products separation. Gels were run at a constant voltage of 100 V for 1 hour, stained in 2 μg/ml ethidium bromide for 10 minutes and photographed under UV by Gel-Document. The expected PCR products for 16S–23S, Capsular K1, K2 and rmpA were 130, 1283, 641 and 537 base pair (bp) in length, respectively.
Results
Serotyping and antimicrobial susceptibility patterns of K. pneumonia
During the study period, a total of 90 K. pneumonia clinical isolates, were collected. The molecular serotyping was performed and showed in Table 4. Among 90 K. pneumonia clinical isolates, 13 had serotype K1A with redundancy of 14.44% and 15 cases had serotype K2A with the redundancy of 16.66%. rmpA gene was observed in 10 isolates the redundancy of 11.11% (Figure 1 and Figure 2). Of the total 90 K. pneumonia clinical isolates, 55 were collected from females and 35 isolates were from males. There was widespread resistance of the isolates to Amoxicillin 87.8%, Cephalothin 53.3%, Kanamycin 45.5%, Tetracycline 43.3%, Ceftriaxon 41.1%, Nitrofurantoin 41.1%, Cotrimoxazole 36.7%, Amikacin 32.2%, Cefepime 34.4%, Gentamicin 26.7% (Table 5).
| Isolate source | 165rRNA | K1A | k2A | RmpA |
|---|---|---|---|---|
| urine (N= 76) | 76 | 11 | 12 | 10 |
| Blood culture (N= 5) | 5 | 2 | 1 | 0 |
| Eye secretion (N= 5) | 5 | 0 | 2 | 0 |
| Wound (N= 2) | 2 | 0 | 0 | 0 |
| Trachea (N= 2) | 2 | 0 | 0 | 0 |
| Total (N= 90) | 90 (100%) | 13 (14.44%) | 15 (16.66%) | 10 (11.11%) |
Table 4: Serotype K1, K2 and rmp isolates from samples.
| Antimicrobial agent | Resistant | Intermediate resistant | Susceptible | |||
|---|---|---|---|---|---|---|
| Number | % | Number | % | Number | % | |
| Amoxicillin | 88 | 97.8 | 1 | 1.1 | 1 | 1.1 |
| Nalidixic acid | 22 | 24.4 | 8 | 9 | 60 | 66.6 |
| Nitrofurantoin | 37 | 41.1 | 22 | 24.5 | 31 | 34.4 |
| Imipenem | 4 | 4.4 | 2 | 2.2 | 84 | 93.4 |
| Cefepime | 31 | 34.4 | 3 | 3.3 | 56 | 62.2 |
| Tetracycline | 39 | 43.3 | 17 | 18.8 | 34 | 37.9 |
| Co-trimoxazole | 33 | 36.7 | 2 | 2.2 | 55 | 61.1 |
| Ciprofloxacin | 6 | 6.7 | 4 | 4.4 | 80 | 88.9 |
| Kanamycin | 41 | 45.5 | 26 | 28.9 | 23 | 25.6 |
| Ceftriaxone | 37 | 41.1 | 5 | 5.5 | 48 | 53.4 |
| Cephalothin | 48 | 53.3 | 6 | 6.7 | 36 | 40 |
| Norfloxacin | 15 | 16.7 | 3 | 3.3 | 72 | 80 |
| Amikacin | 29 | 32.2 | 4 | 4.5 | 57 | 63.3 |
| Gentamicin | 24 | 26.7 | 1 | 1.1 | 65 | 72.2 |
Table 5: Antimicrobial resistance profiles of K. pneumoniae isolates against 90 antimicrobial agents.
Distribution of Klebsiella pneumonia antimicrobial resistance pattern and antibiotic resistance genes
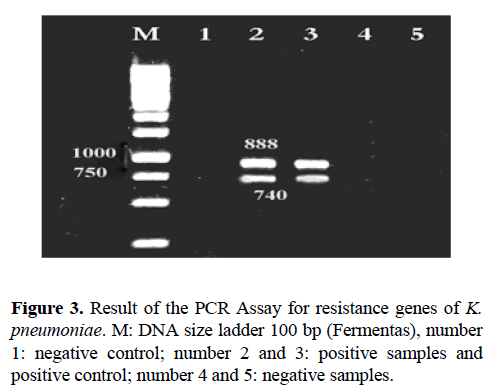
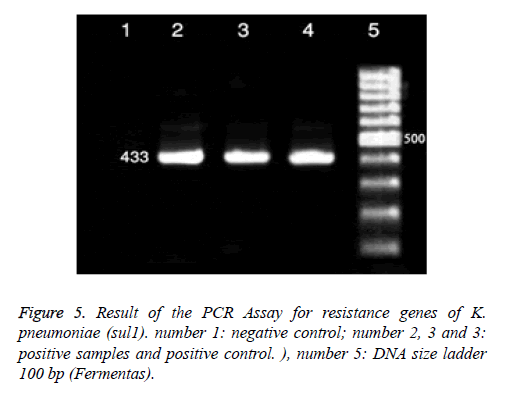
The PCR Assay Result for resistance genes of K. pneumoniae is presented in Figures 3-5 and the frequency of genes reported to tet A 79.48%, tet B 64.10%, sul1 21.21%, aac (3) IIa 83.33%, qnr in the antibiotic nalidixic acid 18.18%, Norfloxacin in three isolates (20%) and ciprofloxacin 16.66% (Table 6). The Antimicrobial resistance pattern of K. pneumoniae isolates are presented in Table 7.
| Gene | Antimicrobial agent | Resistance by disc | Resistance by PCR |
|---|---|---|---|
| tet A | Tetracycline | 39 (43.30%) | 31 (79.48%) |
| tet B | Tetracycline | 39 (43.30%) | 25 (64.10%) |
| Qnr | Nalidixic Acid | 22 (22.40%) | 4 (18.18%) |
| Qnr | Norfeloxacine | 15 (16.70%) | 3 (20%) |
| Qnr | Ciprofloxacin | 6 (6.70%) | 1 (16.16%) |
| sul 1 | Sulfonamide | 33 (36.70%) | 15 (45.45%) |
| aac (3)Iia | Gentamycine | 24 (26.70%) | 20 (83.33%) |
Table 6: Distribution of antibiotic resistance genes in K. pneumoniae strains isolated.
| Isolate | Number of Multidrug-Resistant | Resistance pattern |
|---|---|---|
| 2 | 7 | IPM / FEP / CP/ CRO / AN / GM |
| 3 | 11 | FM , IPM , FEP, TE , SXT, CP , CRO , CF, NOR , AN , GM |
| 4 | 7 | AM , FEP, K , CRO , CF, AN , GM |
| 5 | 4 | AM , TE , SXT, CF, |
| 6 | 9 | FM , AM , FEP, K , CRO , CF, NOR , AN , GM |
| 7 | 6 | FM / AM/ CRO / CF / AN / GM |
| 8 | 9 | K / FEP / FM / AM / CRO / CF / AN / GM / NOR |
| 9 | 1 | NA , IPM , FEP, TE , SXT, CP , K , CRO , CF, NOR , AN , GM |
| 10 | 7 | FM / AM / K/ CRO/ CF / GM / SXT |
| 11 | 5 | FM / AM / CF / TE / NOR |
| 12 | 7 | K / AM / CF / CRO / GM / FEP / AN |
| 13 | 3 | TE / AM / CF |
| 14 | 8 | FM , AM , FEP, TE , K , CRO , CF, NOR , AN , GM |
| 15 | 2 | FM , NA , IPM , FEP, SXT, CP , CRO , NOR , AN , GM |
| 16 | 7 | CF/K/AM/GM/CRO/FEP/AN |
| 17 | 13 | FM, AM , NA , IPM , FEP, TE , CP , K , CRO , CF, NOR , AN , GM |
| 18 | 10 | FM//CF/K/AM/GM/CRO/FEP/AN/TE/NA |
| 19 | 4 | AM/TE/NA/SXT |
| 20 | 3 | AM/FM/SXT |
| 21 | 5 | AM/K/SXT/TE/NA |
| 22 | 8 | FM/AM/SXT/TE/NA/NOR/CF/CRO |
| 23 | 8 | FEP/FM/AM/SXT/TE/NA/CF/CRO |
| 24 | 3 | K/FM/AM |
| 25 | 4 | K/AM/SXT/NA |
| 26 | 7 | K/AM/FEP/CF/CRO/GM/AN |
| 27 | 7 | AM/FEP/CF/CRO/FM/SXT/NA |
| 28 | 7 | AM/FEP/CF/CRO/FM/SXT/NA |
| 29 | 4 | AM/CF/SXT/TE |
| 30 | 8 | AM/CF/K/AN/FEP/CRO/GM/NOR |
| 31 | 9 | K/AM/FM/CF/AN/FEP/CRO/GM/SXT |
| 32 | 8 | K/AM/CF/AN/FEP/CRO/GM/SXT |
| 33 | 7 | K/AM/CF/AN/FEP/CRO/GM |
| 34 | 9 | K/AM/CF/AN/FEP/CRO/GM/TE/NOR |
| 35 | 8 | K/AM/CF/AN/FEP/CRO/GM/SXT |
| 36 | 10 | NA/K/TE/AM/CF/FEP/CRO/NOR/IPM/CP |
| 37 | 9 | NA/K/TE/AM/CF/CRO/NOR/CP/SXT |
| 38 | 4 | AM/NA/K/FM |
| 39 | 4 | AM/K/TE/SXT |
| 40 | 4 | AM/K/TE/FM |
| 41 | 4 | AM/FM/SXT/AN |
| 42 | 3 | TE/AM/FM |
| 43 | 9 | FM/K/AM/AN/CF/CRO/NOR/FEP/GM |
| 44 | 4 | AM/AN/TE/SXT |
| 45 | 6 | TE/CF/AM/AN/SXT/NA |
| 46 | 3 | FM/TE/AM |
| 47 | 4 | TE/AM/NA/SXT |
| 48 | 8 | TE/AM/SXT/K/FM/CRO/CF/FEP |
| 49 | 3 | AM/SXT/CF |
| 50 | 4 | TE/AM/FM/NA |
| 51 | 8 | K/CF/TE/AM/CRO/FEP/AN/GM |
| 52 | 4 | K/AM/AN/FM |
| 53 | 11 | K/AM/NA/CF/TE/CRO/FEP/SXT/IPM/CP/NOR |
| 54 | 8 | AN/K/AM/CF/TE/CRO/FEP/GM |
| 55 | 5 | FM/AM/TE/FEP/NA |
| 56 | 6 | CF/AM/NA/AN/CP/NOR |
| 57 | 4 | TE/AM/FM/SXT |
| 58 | 10 | K/CP/NOR/TE/AM/SXT/CF/NA/CRO/IPM |
| 59 | 6 | K/TE/FM/AM/SXT/NA |
| 60 | 9 | CF/K/TE/FM/AM/AN/FEP/CRO/GM |
| 61 | 5 | CF/TE/FM/AM/SXT |
| 62 | 8 | TE/AM/AN/K/CF/FEP/CRO/GM |
| 63 | 5 | AM/K/CF/FEP/CRO |
| 64 | 5 | AM/K/CF/CRO/SXT |
| 65 | 7 | K/AM/CF/CRO/FEP/AN/GM |
| 66 | 5 | AM/CF/CRO/TE/SXT |
| 67 | 3 | AM/TE/SXT |
| 68 | 3 | AM/FM/SXT |
| 69 | 7 | K/AM/CF/CRO/FEP/AN/GM |
| 70 | 6 | AM/CF/CRO/FEP/FM/SXT |
| 71 | 4 | AM/CF/SXT/TE |
Table 7: Antimicrobial resistance pattern in K. pneumoniae isolates.
Prevalence of sulfonamides resistance-encoding sul genes and their relatedness to class 1 integrons
In this study 33 isolates resistance to cotrimoxazole, aren’t finding sul1 gene in 15 isolated cases, sul2 gene in 20 isolated cases, sul3 gene in 2 isolated cases, sul1 and sul2 gene in 2 isolated cases, sul1 and sul3 gene in 1 isolated cases and sul1 and sul2 and sul3 gene in 15 isolated cases, respectively. Overall the most prevalent sul gene was sul2, found in 20/33 (60.60%) strains, followed by sul1 15/33 (45.45%) and sul3 2/33 (6.06%) (Table 8). Also there were 27 demonstrating int1 genes for cotrimoxazol. The sulfonamides resistance-encoding sul 1 genes in relation to class 1 integrons were found in 22/33 (66.66%) of the K. pneumoniae strains (Table 9).
| No. of isolates with genes | ||||||
|---|---|---|---|---|---|---|
| Strain Characteristics | Sul 1 | Sul 2 | Sul 3 | Sul 1 Sul 2 |
Sul 1 Sul 3 |
Sul 1 Sul 2 Sul 3 |
| Sulfonamide Resistance N=33 |
15 45.45% |
20 60. 60% |
2 6. 06% |
2 6. 06% |
1 3. 03% |
- 0% |
Table 8: Prevalence of sul genes in K. pneumoniae isolates resistant to sulphonamides.
| No. of isolates with genes | |||
|---|---|---|---|
| Strain Characteristics | Sul 1 + Int 1 |
Sul 2 + Int 1 |
Sul 3 + Int 1 |
| Sulfonamide Resistance N=33 |
22 66. 66% |
3 15. 15% |
2 6. 06% |
Table 9: Prevalence of in1 genes in K. pneumoniae isolates resistant to sulphonamides.
Discussion
In this study, we evaluated the antibiotic resistance patterns of K. pneumoniae and the frequency distribution of K. pneumoniae genes and their relatedness with the class 1 integron in K. pneumoniae and sul genes. Integrons have become an important means of horizontal transfer of resistance genes in clinical isolates [22,35]. The present study showed that the most common K. pneumoniae serotype was K2 (15/90; 16.66%), followed by K1 (13/90; 14.44%). magA has been confirmed to be located in the cps (capsular polysaccharide synthesis) gene cluster of serotype K1 of K. pneumoniae and is restricted to serotype K1 isolates, regardless of their sources [36-38]. Our present data show that A total of 10 (11.11%) K. pneumonia isolates carried rmpA which is in contrary with human isolates of K. pneumoniae, in which the rmpA gene is present in both K1 and K2 capsular serotypes, as well as nearly 67% of non–K1/K2 serotypes,17 but the magA gene appears restricted to isolates of the K1 serotype [15]. Therefore, magA is a good tool for molecular typing rather than a major virulence determinant. In contrary, a study conducted in Singapore and Taiwan showed that the most common serotype was K1 (34/73; 46.6%), followed by K2 (15/73; 20.5%). magA was restricted to serotype K1. All K1 or K2 isolates and 66.7% (16/24) of isolates that were neither serotype K1 nor serotype K2 (non-K1/K2) carried rmpA [39]. In addition, another study also showed that Serotype K2 K. pneumoniae is the second most prevalent serotype next to serotype K1 as a cause of pyogenic liver abscess and is also frequently reported in community acquired pneumonia [40].
The treatment of infectious diseases is an important issue for human wellbeing and the daily increase in bacterial resistance has elevated patients’ costs in recent years. In our study, markedly high resistance to Amoxicillin and Cephalothin was noticed in clinical isolates of K. pneumoniae. K. pneumoniae isolates were considerably resistant to cephalosporin has been reported from other parts of the world [41]. Our study, along with other studies, have also demonstrated that the rates of ESBLs production in our country are different from other countries such as; India (57.1%), Turkey (57%) and South Korea (30%), which showed a higher prevalence of ESBL-producing isolates [42-45]. Feizabadi et al. found that the rates of resistance for amikacin, ciprofloxacin, cefepime, ceftazidime, and cefotaxime were; 21.4%, 28%, 76% and 84.0%, respectively [46]. The comparison of our study results with the above-mentioned study shows that antibiotic resistance to four of the previously mentioned antibiotics is higher in our study. In addition, in another study, both non-hospitalized and hospitalized isolates were more resistant to first line drugs including; ampicillin, and trimethoprim-sulfamethoxazole [45]. This result, which is comparable with other studies in developing countries, is due to the widespread use of these drugs because of their low cost and easy administration. Long hospital stay and antibiotic pressure select resistant strains which were colonized in susceptible patients [47]. In these conditions physicians have limited drug choices. High percentage of resistant to Amoxicillin, Cephalothin, Kanamycin and the other beta lactams show the high rate of beta lactam prescription. Also, aminoglycosides are used in combination therapy with beta- lactam antibiotics. Therefore, it is expected to reveal high rate of resistance to aminoglycosides as well as beta-lactams. Although, sequencing analyzes show integrated gene cassettes related to aminoglycoside resistant in most isolates, however, in total there are medium rates of resistance for aminoglycosides (gentamicin, 26.7% and amikacin 32.2%). Our findings with regard to the overall high resistance of K. pneumoniae strains to antibiotics such as Amoxicillin (87.8%), Cephalothin (53.3%), Kanamycin (45.5%), and others studied are in agreement with those of other recent studies [48-51]. This shows the limited possibility of using these antibiotics in the empirical treatment of patients infected with K. pneumonia. Usually sulfonamides resistance is encoded by the sul1, sul2, and sul3 genes. We found that more than half of the K. pneumoniae strains possessed one or more of these sul genes, and in 60.60% of these strains, sulfonamides resistance occurred. This result is in line with others done among E. coli strains, the sul2 gene has been found to be predominant in E. coli strains isolated in UTI episodes [32,52]. In our study, sulfonamides treatment was associated with the occurrence of sul genes and with increased phenotypic resistance to sulfonamides. Horizontal gene transfer has been associated with escalated SXT resistance among Enterobacteriaceae [52,53]. The remarkable stability of resistance markers, such as phenotypic resistance patterns and sul genes, among K. pneumoniae strains may be a helpful tool for the preliminary differentiation between relapse and reinfection.
The present study characterized class 1 integrons and their gene cassettes in K. pneumoniae isolates collected from clinical patients. In this study, we observed lower class 1 integron prevalence in K. pneumoniae (66.66%) compared to the previously reported frequencies of 92% in India [54], 93.2% in Shan Dong, China [55], 73% in Australia [56], and 70% in the United States [25]. The class 1 integron was highly prevalent in K. pneumoniae (66.66%) and was strongly associated with the sul1 genes, which was similar in other literature [57]. Thus, class 1 integrons with various gene cassette arrays in association with sul1 genes were highly prevalent in Enterobacteriaceae, and the variation of the gene cassettes in class 1 integrons may reflect the horizontal transfer of integrons among members of the Enterobacteriaceae family [57]. On the other hand, previous study conducted in Iran showed that Class 1 integrons were more frequent among K. pneumoniae isolates in comparison with class 2. Five different resistance gene arrays were also identified among class 1 integrons. Dihydrofolate reductase (dfrA) and aminoglycoside adenyltransferase (aad) gene cassettes were found to be predominant in the class 1 integrons [58].
In conclusion, we report the first extensive study regarding the distribution and antimicrobial resistant profile of K. pneumoniae and sul genes and the prevalence of sulfonamides resistance-encoding sul genes and their Relatedness to Class 1 Integrons among K. pneumoniae isolates in Iran. The study has revealed that serotype K1 is one of the most important serotypes of K. pneumonia. Also there seems to be a strong relationship between presence of Integron and increased resistance to different antibiotics. In this study serotype K1 or K2 is the major virulence determinant for K. pneumoniae. Majority of the isolates are resistance to Amoxicillin and Cephalothin. In addition, resistance to sulfonamides in K. pneumoniae was explained by the acquisition of sul1, sul2, and sul3 genes. There is also high rate of antibacterial resistance in K. pneumoniae and diverse integrated gene cassettes related to class 1 integrons. In most of the cases, class 1 integrons with various multi-gene cassette arrays in association with sul1 genes were widely disseminated in K. pneumoniae so that, there is a strong relationship between presence of class 1 Integron and increased resistance to sulfonamides antibiotics. The wide distribution of integrons in the K. pneumoniae isolates and sul genes may be because of the horizontal transfer of antibiotic resistance gene and might become a serious threat to the search for effective antimicrobial therapy in the future. The results of this study reinforce the need for increasing concern for therapy for clinical infections caused by K. pneumoniae isolates having resistance-encoding sul genes in relation to class 1 integrons. Therefore, continued monitoring of antimicrobial resistance, the adoption of prudent use of antimicrobial agents and the establishment of a surveillance system is urgently needed to prevent further dissemination in Iran.
References
- Ranjbar R, Izadi M, Hafshejani TT, Khamesipour F. Molecular detection and antimicrobial resistance of Klebsiella pneumoniae from house flies (Musca domestica) in kitchens, farms, hospitals and slaughterhouses. J Infect Public Health 2016.
- Janda JM, Abbott SL. The Genera Klebsiella and Raoultella, pp. 115-135, 3rd ed. In: The Enterobacteria. ASM Press, Washington, D.C 2005.
- Brisse S, Grimont F, Grimont PAD. The Genus Klebsiella. In: Dworkin; 159-196.
- Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. The Prokaryotes: a handbook on the biology of bacteria. Berlin: Springer-Verlag.
- Yulandi A, Sugiokto FG, Febrilina, Suwanto A. Genomic Sequence of Klebsiella pneumoniae IIEMP-3, a Vitamin B12-Producing Strain from Indonesian Tempeh. Genome Announc 2016; 4(1): e01724-15.
- Chang SC, Fang CT, Hsueh PR, Chen YC, Luh KT. Klebsiella pneumoniae isolates causing liver abscess in Taiwan. Diagn Microbiol Infect Dis 2000; 37: 279-284.
- Ko WC, Paterson DL, Sagnimeni AJ, Hansen DS, Von Gottberg A, Mohapatra S, Casellas JM, Goossens H, Mulazimoglu L, Trenholme G. Community acquired Klebsiella pneumoniae bacteremia: global differences in clinical patterns. Emerg Infect Dis 2002; 8: 160-166.
- Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med 2004; 199: 697-705.
- Fang FC, Sandler N, Libby SJ. Liver abscess caused by magA+ Klebsiella pneumoniae in North America. J Clin Microbiol 2005; 43: 991-992.
- Kawai T. Hypermucoviscosity: an extremely sticky phenotype of Klebsiella pneumoniae associated with emerging destructive tissue abscess syndrome. Clin. Infect Dis 2006; 42: 1359-1361.
- Soto E, Marchi S, Beierschmitt A, Kearney M, Francis S, VanNess K, Vandenplas M, Thrall MA, Palmour R. Interaction of non-human primate complement and antibodies with hypermucoviscous Klebsiella pneumoniae. Vet Res 2016; 47: 40.
- Babini GS, Livermore DM. Are SHV beta-lactamases universal in Klebsiella pneumoniae? Antimicrobial Agents and Chemotherapy 2000; 44: 22-30.
- Rawat D, Nair D. Extended-spectrum β-lactamases in Gram Negative Bacteria. J Global Infect Dis 2010; 2: 263-274. Du Bois S, Marriott M, Amyes S. TEM- and SHV derived extended-spectrum β-lactamases: relationship between selection, structure and function. Journal of Antimicrobial Chemotherapy 1995; 35: 7-22.
- Paterson DL, Bonomo RA. Extended-spectrum -lactamases: a clinical update. Clin Microbiol Rev 2005; 18: 657-686.
- Gundogdu A, Long YB, Vollmerhausen TL, Katouli M. Antimicrobial resistance and distribution of sul genes and integron associated intI genes among uropathogenic Escherichia coli in Queensland, Australia. J Med Microbiol 2011; 60: 1633-1642.
- Huovinen P. Resistance to trimethoprim-sulfamethoxazole. Clin Infect Dis 2001; 32: 1608-1614.
- Trobos M, Christensen H, Sunde M, Nordentoft S, Agersø Y, Simonsen GS, Hammerum AM and Olsen JE. Characterization of sulphonamideresistant Escherichia coli using comparison of sul2 gene sequences and multilocus sequence typing. Microbiology 2009; 155: 831-836.
- Skold O. Sulfonamide resistance: mechanisms and trends. Drug Resist 2000; 3: 155-160.
- Toleman MA, Bennett PM, Bennett DM, Jones RN, Walsh TR. Global emergence of trimethoprim/sulfamethoxazole resistance in Stenotrophomonas maltophilia mediated by acquisition of sul genes. Emerg Infect Dis 2007; 13: 559-565.
- Tajbakhsh E, Khamesipour F, Ranjbar R, Ugwu IC. Prevalence of class 1 and 2 integrons in multi-drug resistant Escherichia coli isolated from aquaculture water in Chaharmahal Va Bakhtiari province, Iran. Ann Clin Microbiol Antimicrobials 2015; 14: 37.
- Guo X, Xia R, Han N, Xu H. Genetic diversity analyses of class 1 integrons and their associated antimicrobial resistance genes in Enterobacteriaceae strains recovered from aquatic habitats in China. Lett Appl Microbiol 2011; 52: 667-675.
- Roy C, Ingold A, Vanegas N, Martinez E, Merlino J, Merkier AK, Castro M, Gonzalez RG, Borthagaray G, Centron D. Dissemination of multiple drug resistance genes by class 1 integrons in Klebsiella pneumoniae isolates from four countries: a comparative study. Antimicrob Agents Chemother 2011; 55: 3140-3149.
- Labbate M, Case RJ, Stokes HW. The integron/ gene cassette system: an active player in bacterial adaptation. Methods Mol Biol2009 532: 103-125.
- Kang HY, Jeong YS, Oh JY, Tae SH, Choi CH, Moon DC, Lee WK, Lee YC, Seol SY, Cho DT. Characterization of antimicrobial resistance and class 1 integrons found in Escherichia coli isolates from humans and animals in Korea. J Antimicrob Chemother 2005; 55: 639-644.
- Rao AN, Barlow M, Clark LA, Boring JR, Tenover FC, McGowan JJ. Class 1 integrons in resistant Escherichia coli and Klebsiella spp., US hospitals. Emerg Infect Dis 2006; 12: 1011-1014.
- Seputiené V, Povilonis J, Ruzauskas M, Pavilonis A, Suziedéliené E. Prevalence of trimethoprim resistance genes in Escherichia coli isolates of human and animal origin in Lithuania. J Med Microbiol 2010; 59: 315-322.
- Feizabadi MM, Delfani S, Raji N, Majnooni A, Aligholi M, Shahcheraghi F, Parvin M, Yadegarinia D. Distribution of bla(TEM), bla(SHV), bla(CTX-M) genes among clinical isolates of Klebsiella pneumoniae at Labbafine jad Hospital, Tehran, Iran. Microb Drug Resist 2010; 16: 49-53.
- Turton JF, Perry C, Elgohari S, Hampton CV. PCR characterization and typing of Klebsiella pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets. J Med Microbiol 2010; 59: 541-547.
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing, 20th informational supplement (M100-S20). Wayne Pa: Clinical and Laboratory Standards Institute 2010; 30.
- Sambrook J, Russell DW. Molecular cloning. Laboratory Manual. New York: Cold Spring Harbor Laboratory Press, Cold Spring Harbor. 58-152.
- Maynard C, Bekal S, Sanschagrin F, Levesque RC, Brousseau R, Masson L, Larivière S, Harel J. Heterogeneity among virulence and antimicrobial resistance gene profiles of extraintestinal Escherichia coli isolates of animal and human origin. J Clin Microbiol 2004; 42: 5444-5452.
- Soleimani-Asl Y, Zibaei MF. Detection of qnrA gene among quinolone-resistant Escherichia coli isolated from urinary tract infections in Khorram Abad during 2011-2012. J Kashan Univ Med Sci 2013; 7: 488-494.
- Kerrn MB, Klemmensen T, Frimodt-Moller N, Espersen F. Susceptibility of Danish Escherichia coli strains isolated from urinary tract infections and bacteraemia, and distribution of sul genes conferring sulphonamide resistance. J. Antimicrob. Chemother 2002; 50: 513-516.
- Mazel D. Integrons: agents of bacterial evolution. Nat Rev Microbiol 2006; 4: 608-620.
- Poirel L, Carrer A, Pitout JD, Nordmann P. Integron mobilization unit as a source of mobility of antibiotic resistance genes. Antimicrob Agents Chemother 2009; 53: 2492-2498.
- Struve C, Bojer M, Nielsen EM, Hansen DS, Krogfelt KA. Investigation of the putative virulence gene magA in a worldwide collection of 495 Klebsiella isolates: magA is restricted to the gene cluster of Klebsiella pneumoniae capsule serotype K1. J Med Microbiol 2005; 54: 1111-1113.
- Chuang YP, Fang CT, Lai SY, Chang SC, Wang JT. Genetic determinants of capsular serotype K1 of Klebsiella pneumoniae causing primary pyogenic liver abscess. J Infect Dis 2006; 193: 645-654.
- Yeh KM, Chang FY, Fung CP, Lin JC, Siu LK. magA is not a specific virulence gene for Klebsiella pneumoniae strains causing liver abscess but is part of the capsular polysaccharide gene cluster of K. pneumonia serotype K1. J Med Microbiol 2006; 55: 803-804
- Kuo-Ming Y, Kurup A, Siu LK, Koh YL, Chang-Phone FL, Jung-Chung C, Te-Li C, Feng-Yee, Tse-Hsien K. Capsular Serotype K1 or K2, Rather than magA and rmpA, Is a Major Virulence Determinant for Klebsiella pneumonia Liver Abscess in Singapore and Taiwan. Journal of clinical microbiology 45: 466-471.
- Decre D, Verdet C, Emirian A, Le Gourrierec T, Petit JC, Offenstadt G, Maury E, Brisse S, Arlet G. Emerging severe and fatal infections due to Klebsiella pneumoniae in two university hospitals in France. J Clin Microbiol 2011; 49: 3012-3014.
- Nijssen S, Florijn A, Bonten MJ, Schmitz FJ, Verhoef J, Fluit AC. Beta-lactam susceptibilities and prevalence of ESBL-producing isolates among more than 5000 European Enterobacteriaceae isolates. Int J Antimicrob Agents 2004; 24: 585-591.
- Jeong SH, Bae IK, Lee JH, Sohn SG, Kang GH, Jeon GJ, Kim YH, Jeong BC, Lee SH. Molecular characterization of extended-spectrum beta-lactamases produced by clinical isolates of Klebsiella pneumoniae and Escherichia coli from a Korean nationwide survey. J Clin Microbiol 2004; 42: 2902-2906.
- Tasli H, Bahar IH. Molecular characterization of TEM- and SHV-derived extended-spectrum beta-lactamases in hospital-based Enterobacteriaceae in Turkey. Jpn J Infect Dis 2005; 58: 162-167.
- Lal P, Kapil A, Das BK, Sood S. Occurrence of TEM & SHV gene in extended spectrum beta-lactamases (ESBLs) producing Klebsiella sp. isolated from a tertiary care hospital. Indian J Med Res 2007; 125: 173-178.
- Mobasherizadeh S, Shokri D, Zargarzadeh AH, Jalalpour S, Ebneshahidi SA, Sajadi M. Antimicrobial resistance surveillance among hospitalized and non-hospitalized extend-spectrum beta-lactamase producing Escherichia coli from four tertiary-care hospitals in Isfahan, Iran; 2008-2011. Afr J Microbiol Res 2012; 6: 953-959.
- Feizabadi MM, Mahamadi-Yeganeh S, Mirsalehian A, Mirafshar SM, Mahboobi M, Nili F, Yadegarinia D. Genetic characterization of ESBL producing strains of Klebsiella pneumoniae from Tehran hospitals. J Infect Dev Ctries 2010; 4: 609-615.
- Biedenbach DJ, Moet GJ, Jones RN. Occurrence and antimicrobial resistance pattern comparisons among bloodstream infection isolates from Sentry Antimicrobial Surveillance Program. Diagn Microbiol Infect Dis 2004; 50: 59-69.
- Karki T, Truusalu K, Vainumae I, Mikelsaar M. Antibiotic susceptibility patterns of community- and hospital-acquired Staphylococcus aureus and Escherichia coli in Estonia. Scand J Infect Dis 2001; 33: 333-338.
- Anatoliotaki M, Galanakis E, Schinaki A, Stefanaki S, Mavrokosta M, Tsilimigaki A. Antimicrobial resistance of urinary tract pathogens in children in Crete, Greece. Scand. J Infect Dis 2007; 39: 671-675.
- Garcia Garcia MI, Munoz Bellido JL, Garcia Rodriguez JA. In vitro susceptibility of community-acquired urinary tract pathogens to commonly used antimicrobial agents in Spain: a comparative multicenter study (2002–2004). J Chemother 2007; 19: 263-270.
- Prelog M, Schiefecker D, Fille M, Wurzner R, Brunner A, Zimmerhackl LB. Febrile urinary tract infection in children: ampicillin and trimethoprim insufficient as empirical mono-therapy. Pediatr Nephrol 2008; 23: 597-602.
- Blahna MT, Zalewski CA, Reuer J, Kahlmeter G, Foxman B, Marrs CF. The role of horizontal gene transfer in the spread of trimethoprim- sulfamethoxazole resistance among uropathogenic Escherichia coli in Europe and Canada. J Antimicrob Chemother 57: 666-672.
- Peirano G, Agerso Y, Aarestrup FM, dos Reis EM, dos Prazeres Rodrigues D. Occurrence of integrons and antimicrobial resistance genes among Salmonella enterica from Brazil. J Antimicrob Chemother 2006; 58: 305-309.
- Bhattacharjee A, Sen MR, Prakash P, Gaur A, Anupurba S, Nath G. Observation on integron carriage among clinical isolates of Klebsiella pneumoniae producing extended-spectrum beta-lactamases. Indian J Med Microbiol 2010; 28: 207-210.
- Yao F, Qian Y, Chen S, Wang P, Huang Y. Incidence of extended-spectrum beta-lactamases and characterization of integrons in extended-spectrum beta-lactamase producing Klebsiella pneumoniae isolated in Shantou, China. Acta Biochim Biophys Sin (Shanghai) 2007; 39: 527-532.
- Jones LA, McIver CJ, Kim MJ, Rawlinson WD, White PA. The aad B gene cassette is associated with blaSHV genes in Klebsiella species producing extended spectrum beta-lactamases. Antimicrob Agents Chemother 2005; 49: 794-797.
- Li B, Hu Y, Wang Q, Yi Y, Woo PC, Jing H, Zhu B, Liu CH. Structural diversity of class 1 integrons and their associated gene cassettes in Klebsiella pneumoniae isolates from a hospital in China. PLoS One 2013; 30: e75805.
- Zeighami H, Haghi F, Hajiahmadi F. Molecular characterization of integrons in clinical isolates of betalactamase-producing Escherichia coli and Klebsiella pneumoniae in Iran. J Chemother 2015; 27: 145-151.