ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 10
Analysis of mitochondrial tRNA mutations in patients with acute myocardial infarction
Ren-Shu Wang, Shu-Li Liu, Kai-Hui Zheng and Rong-Kai You*
Department of Intensive Care Unit, Wenzhou Central Hospital, Wenzhou Medical University, PR China
- *Corresponding Author:
- Rong-Kai You
Department of Intensive Care Unit
Wenzhou Central Hospital
Wenzhou Medical University
PR China
Accepted date: February 17, 2017
Background: Mutations in mitochondrial DNA (mtDNA) were the important causes of cardiovascular diseases. However, little was known regarding the role of mitochondrial tRNA (mt-tRNA) mutations in acute Myocardial Infarction (AMI).
Objective: To investigate the association between mt-tRNA mutations and AMI in Han Chinese population.
Methods: 100 unrelated AMI patients and 50 control subjects were recruited in this study; we performed PCR-RFLP to amplify 22 mt-tRNA genes and subsequently sequenced the PCR products. In addition, the pathogenicity scoring system was used to evaluate the deleterious roles of these mt-tRNA mutations. We also used real-time PCR method to determine the mtDNA content in AMI patients carrying these tRNA mutations.
Results: 4 tRNA mutations were identified by PCR-Sanger sequencing; these mutations included tRNAIle A4295G, tRNAMet A4435G, tRNAAla T5655C, tRNALeu (CUN) A12308G. Notably, these mutations were localized at the highly conserved nucleotides of the corresponding tRNAs, may cause the failure in mt-tRNA metabolism. Moreover, AMI group showed a marked reduction in mitochondrial copy number when compared with the controls (P<0.05).
Conclusions: Mutations in mt-tRNA genes were the important causes of AMI, our findings provided novel insight into the pathophysiology of AMI that were manifested by mitochondrial dysfunction.
Keywords
Mt-tRNA, Mutations, AMI, Copy number, Mitochondrial dysfunction
Introduction
Coronary Heart Disease (CHD) is the leading cause of death in the world and accounted for 7.4 million deaths in 2012 [1]. In Europe the mortality per annum from CHD is 20% [2] and 25% in the United States [3]. Acute Myocardial Infarction (AMI), a common type of CHD, is the leading cause of morbidity and mortality worldwide [4,5], governments and patients have to carry big economic burden in both developed and developing countries from the disease itself and post-infarction heart failure in this post-drug eluting stenting era [6]. Thus, an effective and accurate diagnostic biomarker would be needed to help decrease the mortality of AMI patients.
Mitochondrion is a unique cell organelle, playing an important role in production of ATP by Oxidative Phosphorylation (OXPHOS) [7]. Mitochondrial dysfunctions caused by mtDNA mutations have been found to be associated with a large number of human diseases, such as neurodegenerative diseases, cardiovascular disorders, cancer and obesity [8]. Among these, point mutations in mitochondrial tRNA (mt-tRNA), were the most common genetic causes of CHD, recently, several mt-tRNA mutations have been reported to be associated with CHD, these mutations included tRNALeu (CUN) A12330G [9], tRNAMet and tRNAGln A4401G [10], tRNAIle A4263G [11]. However, the relationship between mt-tRNA mutations and AMI remained poorly understood.
With the purpose of understanding the role of mitochondrial mutations in AMI, we recently carried out a systematic mutational analysis of mt-tRNA genes in 100 patients with AMI and 50 healthy subjects. Moreover, to see the contribution of mt-tRNA mutations to mitochondrial dysfunction, we evaluated the mtDNA content for these AMI patients carrying the mt-tRNA mutations.
Materials and Methods
Subjects
A total of 100 genetically unrelated Han Chinese subjects with AMI (45% males and 55% females, aged 44 to 68 years) were recruited between June 2015 and June 2016 at Wenzhou Central Hospital, affiliated to Wenzhou Medical University. Moreover, 50 healthy controls with the age- and gender-matched were collected in the same area. Informed consent, blood samples and clinical evaluation were obtained from all of the participants, approved by the Ethics Committee of the Wenzhou Central Hospital, affiliated to Wenzhou Medical University.
AMI was defined based on elevated cardiac troponin-I or T level (exceeding upper limit of normal) or creatine Kinase-MB fraction (CK-MB) (exceeding three times upper limit of normal), along with angiographic evidence. Angiographic evidence for AMI included significant coronary stenosis, i.e., more than 50% luminal stenosis, intracoronary filling defect or haziness suggesting coronary thrombus/vulnerable plaque, or coronary artery vasospasm confirmed by intracoronary acetylcholine or ergonovine provocation test.
DNA extraction, PCR amplification and sequencing
Genomic DNA was isolated from the whole blood of participants using a TaKaRa Blood Genome DNA Extraction Kit (TaKaRa Biotechnology). In addition, we used the primers for genetic amplification of the 22 mt-tRNA genes, the information of primers were listed in Table 1, PCR mixture included 200 μm dNTP, 10X buffer, Taq DNA polymerase and 15 mmol/L Mg2+ (TaKaRa Biotechnology). Each fragment was purified and subsequently analysed by direct sequencing in an ABI 3700 automated DNA sequencer using the Big Dye Terminator Cycle sequencing reaction kit (Version 3.1). These sequence results were compared with the updated consensus Cambridge sequence to identify the nucleotides alternations (GenBank accession number: NC_012920) [12].
| Target gene | Primer name | Primer Sequence (5’-3’) | Product size |
|---|---|---|---|
| tRNAPhe | MT-1F | CTCCTCAAAGCAATACACTG | 802 bp |
| MT-1R | TGCTAAATCCACCTTCGACC | ||
| tRNAVal | MT-2F | CGATCAACCTCACCACCTCT | 802 bp |
| MT-2R | TGGACAACCAGCTATCACCA | ||
| tRNALeu (UUR) | MT-4F | AAATCTTACCCCGCCTGTTT | 887 bp |
| MT-4R | AGGAATGCCATTGCGATTAG | ||
| tRNAIle | MT-6F | TGG CTC CTT TAA CCT CTC CA | 898 bp |
| tRNAGln | |||
| tRNAMet | MT-6R | AAG GAT TAT GGA TGC GGT TG | |
| tRNAAla | MT-8F | CTAACCGGCTTTTTGCCC | 814 bp |
| tRNAAsn | |||
| tRNACys | MT-8R | ACCTAGAAGGTTGCCTGGCT | |
| tRNASer (UCN) | MT-11F | ACGCCAAAATCCATTTCACT | 987 bp |
| tRNAAsp | MT-11R | CGGGAATTGCATCTGTTTTT | |
| tRNALys | MT-12F | ACG AGT ACA CCG ACT ACG GC | 900 bp |
| MT-12R | TGG GTG GTT GGT GTA AAT GA | ||
| tRNAGly | MT-15F | TCTCCATCTATTGATGAGGGTCT | 891 bp |
| tRNAArg | MT-15R | AATTAGGCTGTGGGTGGTTG | |
| tRNAHis | MT-18F | TATCACTCTCCTACTTACAG | 866 bp |
| tRNASer(AGY) | |||
| tRNALeu (CUN) | MT-18R | AGAAGGTTATAATTCCTACG | |
| tRNAGlu | MT-21F | GCATAATTAAACTTTACTTC | 938 bp |
| MT-21R | AGAATATTGAGGCGCCATTG | ||
| tRNAThr | MT-22F | TGAAACTTCGGCTCACTCCT | 1162 bp |
| tRNAPro | MT-22R | GAGTGGTTAATAGGGTGATAG |
Table 1: Primers for PCR amplification of the mt-tRNA genes.
Phylogenetic conservation analysis
For inter-specific analysis of those variants identified, a total of 17 mitochondrial sequences were used as described elsewhere [13]. The Conservation Index (CI) was calculated by comparing the human mtDNA variants with other 16 vertebrates. Notably, the CI ≥ 70% was regarded as having functional potential [14].
Qualification of mtDNA copy number
After PCR-RFLP, we selected the samples carrying tRNA mutations and 5 controls for further mtDNA content analysis. The relative mtDNA copy number was measured by a realtime PCR and corrected by simultaneous measurement of the nuclear DNA according to the method developed by Wong and Cortopassi [15]. Essentially, measurements were carried out using the Light Cycler-Fast Start DNA Master SYBR Green I kit supplied by Roche Molecular Biochemicals (Pleasanton, CA). The mtDNA content was normalized to a single copy nuclear β-globin gene. Two primers (forward: 5’- GAAGAGCCAAGGACAGGTAC-3’, reverse: 5’- CAACTTCATCCACGTTCACC-3’) complementary to the sequence of β-globin gene were used to amplify a 268-bp product. To amplify the mitochondrial genome, (forward: 5’- AACATACCCATGGCCAACCT-3’ and the reverse: 5’- AGCGAAGGGTTGTAGTAGCCC-3’), which were complementary to the sequence of the mitochondrial ND1 gene, were used to amplify a 153-bp PCR product. We first generated standard curves for both fragments and calculated their respective amplification efficiencies to test if using the 2- CT method was appropriate. All samples were assayed in duplicate.
Assigning pathogenicity to mt-tRNA mutations
The pathogenicity classification of these identified mt-tRNA mutations were assigned using an updating version of a previous validated scoring system [16]. This pathogenicity scoring system employed a number of weighted criteria covering a range of molecular and genetic data, from which an overall pathogenicity score can be obtained. In particular, a variant was classified as “definitely pathogenic” with a score≥ 11 points, whereas a variant was defined as “possible pathogenic” with a core of 7 to 10 points and a “neutral polymorphism” with a score of ≤ 6 points.
Statistically analysis
Data are described as the mean ± SD, statistical significant was evaluated by a one-way ANOVA or independent Student’s t test using SPSS18.0 software (IBM, Armonk), the P<0.05 was considered as statistical significant.
Results
Mutational analysis of mt-tRNA genes
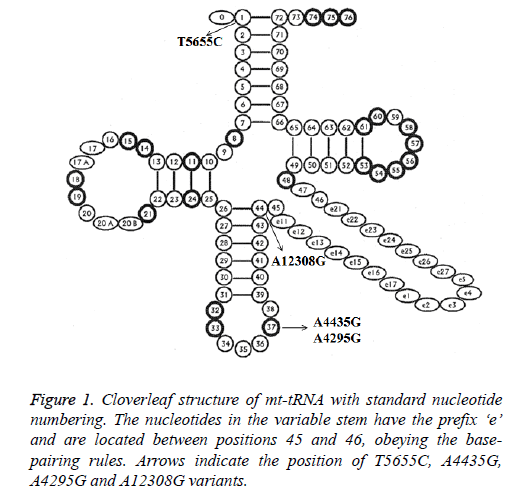
Analysis of the complete mt-tRNA genes in patients with AMI led us to identify 4 mutations: tRNAIle A4295G, tRNAMet A4435G, tRNAAla T5655C and tRNALeu (CUN) A12308G (Table 2). Of these, the A4295G mutations was detected 2 out of 100 AMI patients (2%), the A4435G mutation was found in 1 patient (1%), T5655C mutation was detected in 1 patient (1%) and the A12308G mutation was detected in 2 patients (2%). However, we failed to detect any mt-tRNA mutations in control subjects, in addition, all these mutations were not identified in healthy controls; the location of each mt-tRNA mutation was displayed in Figure 1.
| tRNA species | Position | Replacement | Conservation Index (CI) (%)a | Location of structure | Numbering of tRNA nucleotidesb | No. of 100 patients (%) | No. of 50 controls (%) | Previously reportedc |
|---|---|---|---|---|---|---|---|---|
| tRNAIle | 4295 | A to G | 98.1 | Anticodon stem | 37 | 2 (0.2) | 0 | Yes |
| tRNAMet | 4435 | A to G | 100 | Anticodon stem | 37 | 1 (0.1) | 0 | Yes |
| tRNAAla | 5655 | T to C | 71.1 | Acceptor arm | 1 | 1 (0.1) | 0 | Yes |
| tRNALeu (CUN) | 12308 | A to G | 98.1 | Variable region | 44 | 2 (0.2) | 0 | Yes |
| aConservation index indicated the conservative properties of the nucleotides in 17 species. bNumbers represent the nucleotide positions according to the mitotRNAdb http://mttrna.bioinf.uni-leipzig.de/mtDataOutput. cAccording to MITOMAP (http://www.mitomap.org/MITOMAP). |
||||||||
Table 2: Molecular characterization of mt-tRNA variants associated with AMI.
Pathogenicity scoring system for tRNA mutations
According to the revised pathogenicity scoring system [16], we noticed that the total scores of A4295G, A4435G, T5655C and A12308G mutations were 17, 13, 11 and 15 points, respectively (Table 3), suggesting that all of them should be regarded as “definitely pathogenic”.
| Scoring criteria | A4295G variant | Score/20 | A4435G variant | Score/20 | T5655C variant | Score/20 | A12308G variant | Score/20 | Classification |
|---|---|---|---|---|---|---|---|---|---|
| More than one independent report | Yes | 2 | Yes | 2 | Yes | 2 | Yes | 2 | ≤ 6 points: neutral polymorphisms; 7~10 points: possibly pathogenic; ≥ 11 points: definitely pathogenic |
| Evolutionary conservation of the base pair | 2 | No changes | 2 | two changes | 0 | No changes | 2 | ||
| Variant heteroplasmy | No | 0 | No | 0 | No | 0 | No | 0 | |
| Segregation of the mutation with disease | Yes | 2 | Yes | 2 | Yes | 2 | Yes | 2 | |
| Histochemical evidence of mitochondrial disease | Strong evidence | 2 | No evidence | 0 | No evidence | 0 | Yes | 2 | |
| Biochemical defect in complex I, III or IV | Yes | 2 | Yes | 2 | Yes | 2 | Yes | 2 | |
| Evidence of mutation segregation with biochemical defect from single-fiber studies | Yes | 2 | No | 0 | No | 0 | No | 0 | |
| Mutant mt-tRNA steady-state level or evidence of pathogenicity in trans-mitochondrial cybrid studies | Yes | 5 | Yes | 5 | Yes | 5 | Yes | 5 | |
| Maximum score | definitely pathogenic | 17 | definitely pathogenic | 13 | definitely pathogenic | 11 | definitely pathogenic | 15 |
Table 3: The pathogenicity scoring system for the mt-tRNA variants.
MtDNA copy number analysis
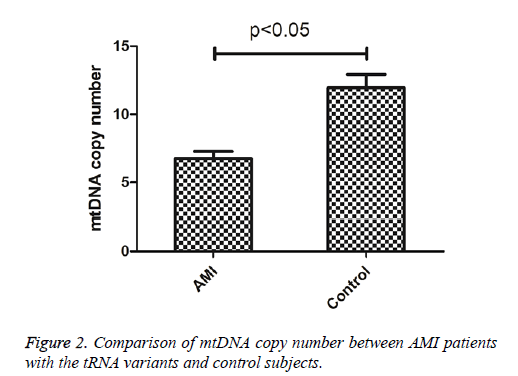
To see whether mutations in tRNA genes caused the alternation of mitochondrial function, the mtDNA content of AMI patients carrying tRNA mutations and control subjects were determined. As shown in Figure 2, the mtDNA copy number was significantly lower than these control subjects (P<0.05), indicating that mutations in tRNA genes may cause mitochondrial dysfunction responsible for AMI.
Discussion
In the present study, we investigated the possible roles of mttRNA mutations in patients with AMI. To the best of our knowledge, this is the first report regarding the association between mt-tRNA mutations and AMI. Recently, increasing evidence showed that mitochondrial dysfunction played important roles in the pathogenesis of cardiovascular diseases [17]. Dysfunction of mitochondrial activity was frequently associated with cardiovascular disease such as heart failure [18]. Today, approximately 200 pathogenic mutations had been mapped to mt-tRNA genes [19], emphasizing the importance of mt-tRNAs for mitochondrial function.
In the current study, we identified 4 mt-tRNA mutations using PCR-Sanger sequencing. Among these, the A4295G and A4435G mutations were localized at 3’end adjacent to the anticodon (position 37) of tRNAIle and tRNAMet, respectively [20]. The adenine at this position of tRNAs was extraordinarily conserved from bacteria to human mitochondria [20]. Almost all of A37 in tRNAs were modified, thus, contributed to the high fidelity of codon recognition, the structural formation and stabilization of functional tRNAs [21]. Notably, the homoplasmic A4295G mutation reduced the efficiency with which tRNAIle can be processed by 3’-tRNase, decreased the level of tRNAIle, thereby led to the defects in mitochondrial translation [22]. Moreover, the A4435G mutation had been reported to increase the penetrance and expressivity of Leber’s Hereditary Optic Neuropathy (LHON)-associated ND4 G11778A mutation in a Han Chinese family [23], approximately~50% reduction in the steady-state level of tRNAMet was observed in the cell lines derived from the individuals carrying the A4435G mutation [23]. While the T5655C mutation in tRNAAla gene was first described in an African family with non-syndromic hearing impairment [24], this mutation disrupted the highly conserved base pairing (A1- U72) at the acceptor arm of tRNAAla, a ~41% reduction in the steady-state level of tRNAAla in mutant cybrids was found, in addition, marked decreases in the level of mitochondrial ATP and membrane potential were also observed in mutant cells with the T5655C mutation [25]. Furthermore, the A12308G mutation had been regarded as a synergistic effect with the mtDNA T12297C mutation associated with dilated cardiomyopathy [26]. Previous study showed that patients with this mutation had a higher relative risk of developing pigmentary retinopathy, short stature, dysphasia-dysarthria and cardiac conduction defects [27], suggesting that this mutation was involved in the development of AMI.
We further investigated whether mutations in tRNA genes caused the mitochondrial dysfunction, for this purpose, the mtDNA copy numbers in peripheral blood of AMI patients carrying these mutations and the healthy controls were determined. We found that mtDNA content was significantly lower in AMI patients when compared with the controls (P<0.05). The mtDNA copy number was a relative measure of the cellular number or mass of mitochondria. Recent experimental studies suggested that alternations in mtDNA copy number played a fundamental role in the increase in ROS, maintenance of mtDNA content was essential for the preservation of mitochondrial function and cell growth [28]. Therefore, we proposed that mt-tRNA mutations may reduce the mtDNA copy number and subsequently result the failure in mitochondrial translation. As a result, dysfunction in the mitochondrial activity may be involved in the development of AMI.
Based on these observations, we proposed that the possible molecular mechanism underlying mt-tRNA mutations in the pathogenesis of AMI may be as follows; first of all, mutations disrupted the secondary structure of mt-tRNA and subsequently resulted the failure in tRNA metabolism such as aminoacylation and post-transcriptional modification [29]. Whatever the consequence may be, the expected net effect would be a decrease in mitochondrial protein synthesis. Defects in mitochondrial translation consequently led to a respiratory phenotype and a decline in ATP production below the threshold level required for normal cell function, thereby causing the mitochondrial dysfunction and longstanding increase of ROS in cardiovascular cells. Thus, our findings provided novel insight into the understanding of pathophysiology and valuable information for management and treatment of AMI.
Conflict of Interest
None
References
- Silva H, Freitas J, Moreira S, Santos A, Almeida V. Alexithymia and psychopathology in patients with acute myocardial infarction. Acta Cardiol 2016; 71: 213-220.
- Nichols M, Townsend N, Scarborough P, Rayner M. Cardiovascular disease in Europe 2014: epidemiological update. Eur Heart J 2014; 35: 2929.
- Minino AM, Murphy SL, Xu J, Kochanek KD. Deaths: final data for 2008. Natl Vital Stat Rep 2011; 59: 1-126.
- Thygesen K, Alpert JS, Jaffe AS, Simoons ML, Chaitman BR, White HD. Third universal definition of myocardial infarction. J Am Coll Cardiol 2012; 60: 1581-1598.
- Xu F, Ming Q, Hou L. The effect of sex counselling in the sexual activity of acute myocardial infarction patients after primary percutaneous coronary intervention. Acta Cardiol 2015; 70: 460-464.
- Lee S, Baek K, Chun K. Cost-effectiveness of drug-eluting vs.bare-metal stents in patients with coronary artery disease from the Korean National Health Insurance Database. Yonsei Med J 2014; 55: 1533-1541.
- Finsterer J, Stollberger C, Haberler C. Dilated cardiomyopathy and recurrent myorrhexis suggest mitochondrial disorder. Acta Cardiol 2016; 71: 491-492.
- Greaves LC, Reeve AK, Taylor RW, Turnbull DM. Mitochondrial DNA and disease. J Pathol 2012; 226: 274-286.
- Teng L, Zheng J, Leng J, Ding Y. Clinical and molecular characterization of a Han Chinese family with high penetrance of essential hypertension. Mitochondrial DNA 2012; 23: 461-465.
- Li R, Liu Y, Li Z, Yang L, Wang S, Guan MX. Failures in mitochondrial tRNAMet and tRNAGln metabolism caused by the novel 4401A>G mutation are involved in essential hypertension in a Han Chinese Family. Hypertension 2009; 54: 329-337.
- Chen X, Zhang Y, Xu B, Cai Z, Wang L, Tian J, Liu Y, Li Y. The mitochondrial calcium uniporter is involved in mitochondrial calcium cycle dysfunction: Underlying mechanism of hypertension associated with mitochondrial tRNA(Ile) A4263G mutation. Int J Biochem Cell Biol 2016; 78: 307-314.
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 1999; 23: 147.
- Lu J, Li Z, Zhu Y, Yang A, Li R, Zheng J, Cai Q, Peng G, Zheng W, Tang X, Chen B, Chen J, Liao Z, Yang L, Li Y, You J, Ding Y, Yu H, Wang J, Sun D, Zhao J, Xue L, Wang J, Guan MX. Mitochondrial 12S rRNA variants in 1642 Han Chinese pediatric subjects with aminoglycoside-induced and non-syndromic hearing loss. Mitochondrion 2010; 10: 380-390.
- Ruiz-Pesini E, Wallace DC. Evidence for adaptive selection acting on the tRNA and rRNA genes of human mitochondrial DNA. Hum Mutat 2006; 27: 1072-1081.
- Wong A, Cortopassi G. Reproducible quantitative PCR of mitochondrial and nuclear DNA copy number using the LightCycler. Methods Mol Biol 2002; 197: 129-137.
- Yarham JW, Al-Dosary M, Blakely EL, Alston CL, Taylor RW. A comparative analysis approach to determining the pathogenicity of mitochondrial tRNA mutations. Hum Mutat 2011; 32: 1319-1325.
- Han JS, Wang HS, Yan DM, Wang ZW, Han HG. Myocardial ischaemic and diazoxide preconditioning both increase PGC-1 alpha and reduce mitochondrial damage. Acta Cardiol 2010; 65: 639-644.
- Urbonavicius S, Wiggers H, Botker HE, Nielsen TT, Kimose HH, Ostergaard M, Lindholt JS, Vorum H, Honore B. Proteomic analysis identifies mitochondrial metabolic enzymes as major discriminators between different stages of the failing human myocardium. Acta Cardiol 2009; 64: 511-522.
- Ruiz-Pesini E, Lott MT, Procaccio V, Poole JC, Brandon MC, Mishmar D, Yi C, Kreuziger J, Baldi P, Wallace DC. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res 2007; 35: 823-828.
- Florentz C, Sohm B, Tryoen-Toth P, Putz J, Sissler M. Human mitochondrial tRNAs in health and disease. Cell Mol Life Sci 2003; 60: 1356-1375.
- Haag S, Sloan KE, Ranjan N, Warda AS, Kretschmer J, Blessing C, Hübner B, Seikowski J, Dennerlein S, Rehling P, Rodnina MV, Hobartner C, Bohnsack MT. NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. EMBO J 2016; 35: 2104-2019.
- Levinger L, Giege R, Florentz C. Pathology-related substitutions in human mitochondrial tRNA (Ile) reduces precursor 3 end processing efficiency in vitro. Nucleic Acids Res 2003; 31: 1904-1912.
- Qu J, Li R, Zhou X, Tong Y, Lu F. The novel A4435G mutation in the mitochondrial tRNAMet may modulate the phenotypic expression of the LHON-associated ND4 G11778A mutation. Invest Ophthalmol Vis Sci 2006; 47: 475-483.
- Li X, Fischel-Ghodsian N, Schwartz F, Yan Q, Friedman RA, Guan MX. Biochemical characterization of the mitochondrial tRNASer (UCN) T7511C mutation associated with nonsyndromic deafness. Nucleic Acids Res 2004; 32: 867-877.
- Jiang P, Wang M, Xue L, Xiao Y, Yu J, Wang H, Yao J, Liu H, Peng Y, Liu H, Li H, Chen Y, Guan MX. A hypertension-associated tRNAAla mutation alters tRNA metabolism and mitochondrial function. Mol Cell Biol 2016; 36: 1920-1930.
- Grasso M, Diegoli M, Brega A, Campana C, Tavazzi L, Arbustini E. The mitochondrial DNA mutation T12297C affects a highly conserved nucleotide of tRNA(Leu(CUN)) and is associated with dilated cardiomyopathy. Eur J Hum Genet 2001; 9: 311-315.
- Crimi M, Del Bo R, Galbiati S, Sciacco M, Bordoni A, Bresolin N, Comi GP. Mitochondrial A12308G polymorphism affects clinical features in patients with single mtDNA macrodeletion. Eur J Hum Genet 2003; 11: 896-898.
- Jeng JY, Yeh TS, Lee JW, Lin SH, Fong TH. Maintenance of mitochondrial DNA copy number and expression are essential for preservation of mitochondrial function and cell growth. J Cell Biochem 2008; 103: 347-357.
- Brule H, Holmes WM, Keith G, Giege R, Florentz C. Effect of a mutation in the anticodon of human mitochondrial tRNAPro on its post-transcriptional modification pattern. Nucleic Acids Res 1998; 26: 537-543.