ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2016) Computational Life Sciences and Smarter Technological Advancement
An optimized approach for enhancement of medical images
1Department of Computer Engineering, Padre Conceicao College of Engineering, Goa, India
2Faculty of Computing, Botho University-Gaborone, Botswana
3Faculty of Engineering, Botho University-Gaborone, Botswana
- *Corresponding Author:
- Meenakshi Sundaram K
Faculty of Computing, Botho University-Gaborone, Botswana
Accepted on September 7, 2016
Ultrasound, an imaging technique based on high-frequency sound waves has been of great importance to the diagnostician and to the medical domain as a whole. Applications of ultrasound have grown tremendously. Furthermore advancements in technology developed three-dimensional (3D) ultrasound that provided more efficient evaluation and diagnosis of the anatomical structure by creating a virtual reality for the viewer. Complex nature of the images makes analysis difficult for the doctor. Apart from the various advantages of being non-invasive, radiation free and cheap a major drawback of ultrasound images is the presence of speckle noise, hindering accurate analysis. A new approach for enhanced ultrasound analysis is put forth considering an additional parameter of intensity in the filtering and clustering process. Visual inspection is considered to be the best testing tool.
Keywords
Filtering, Image enhancement, Segmentation, Ultrasound.
Introduction
Ultrasound being a non-invasive, radiation free and a cheap technique for medical image analysis, it has been very beneficial in providing qualitative and lifesaving information about the internal organs of the human system. It is widely used to observe the conditions related to abdominal tissues and organs, Bone sonometry, foetal monitoring, breast ultrasound, echocardiograms and much more.
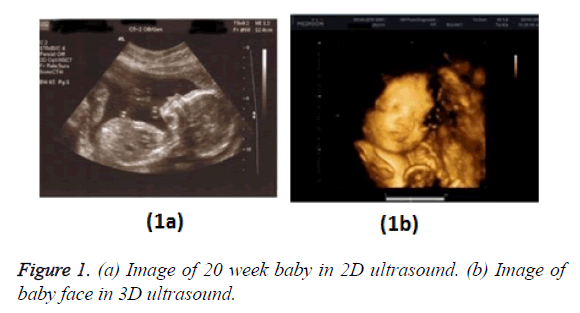
Incorporating the 3D image technology into the ultrasound method was a major advancement in the medical domain allowing the diagnostician to perform accurate and efficient diagnoses as compared to 2D or B-mode ultrasound. In 3D ultrasound the 3D image of the interested object is created by digitally adding together many 2D planes and using the pixel values to determine the voxel value of the 3D image. Huge generation of images has rapidly increased the medical data set which is difficult to handle manually moreover due to the complex internal structure of the human body interpretation and analysis of ultrasound is highly limited hence image segmentation becomes extremely necessary.
Segmentation digitally partitions an image into multiple segments to simplify and change the representation of a complex image into one that is easier to analyse and process. Extending the basic concept of image segmentationsegmentation on 3D medical images can be termed as volume segmentation. 3D image analysis in medical applications is represented by voxels i.e. a value of the image in threedimensional space. Voxel is a coined term of “volume” and “pixels”.
3D Ultrasound images can be acquired in 3 ways [1]:
1. Using mechanically swept probes
2. Using 2D transducer array
3. Freehand acquisition
Figure 1 shows a comparison of B-mode (2D) and 3D foetal ultrasound [2].
Every coin has two sides; ultrasound too has its drawbacks.
Ultrasound images are corrupted by noise speckles; they are affected by shadows, attenuation and missing boundary making analysis difficult and challenging. Thus a major task is to enhance the ultrasound image by de-noising the speckle noise present in the image. Pre-processing of ultrasound image is an essential step to be carried out before using the image for further processing.
Technique
An image obtained is not always 100% pure; it is corrupted by additional unwanted signals which are termed as noise. Noise in an image is introduced during the acquisition and transmission of the image. Noise can be classified into different categories: impulse noise, amplifier noise, multiplicative noise, periodic noise. Based on the category the noise belongs specific de-noising approaches are adopted to handle the removal of noise.
Ultrasound imaging is a very important diagnostic tool in medical analysis. Unfortunately these images are degraded by the presence of speckle noise which affects the fine details thus hindering the accurate analysis of the structure.
Speckle noise reduction is a mandatory pre-processing step for effective diagnosis [3]. During the removal of noise it is necessary to make sure the essentials features of the image are preserved and not lost during de-noising. A key property of speckle noise is its multiplicative nature.
Speckle noise can be represented as [4] shown in Equation 1
G (n, m) = f (n, m)* u (n, m) + ξ (n, m) → (1)
where g (n, m) is the observed image, u (n, m) is the multiplicative component and ξ (n, m) is the additive component of the speckle noise. Here n and m denotes the axial and lateral indices of the image samples.
However, Equation 1 consists of an additive component
ξ (n, m) which does not play a major role in the computation hence this additive component for ultrasound images is almost negligible and can be ignored. Thus Equation 1 can be rewritten as shown in Equation 2
g (n, m) =f (n, m) × u (n, m) → (2)
A convenient approach to remove noise from an image is by the technique of filtering. Filtering of an image is usually performed to either smoothen the image by suppressing high frequencies or enhance the image by suppressing low frequencies. Among the various available filters it has been seen that the median filter is the most appropriate [5,6] for speckle noise removal in ultrasound images. Median filter is a non-linear filter widely used as it is very effective for removing noise while preserving edges. It works by moving through the image pixel by pixel, replacing each value with the median value of neighbouring pixels.
Processing the image by only filtering gives an output fair enough but not very satisfactory thus in this study we put forth a new approach considering additional parameters along with the technique of segmentation. The outcome of the initial approach leads us to incorporate additional parameters to yield improved outcomes in the next approaches. Details and analysis of the approaches are given below.
Data set
The data used in this study is an ultrasound image of a 29 week foetus received from Apollo Victor Hospital Margao, Goa, a franchisee of Apollo Hospitals Enterprise LTD with the consultation of Dr. Paresh Desai, Senior Consultant and Radiologist at Apollo Victor, Margao, Goa.
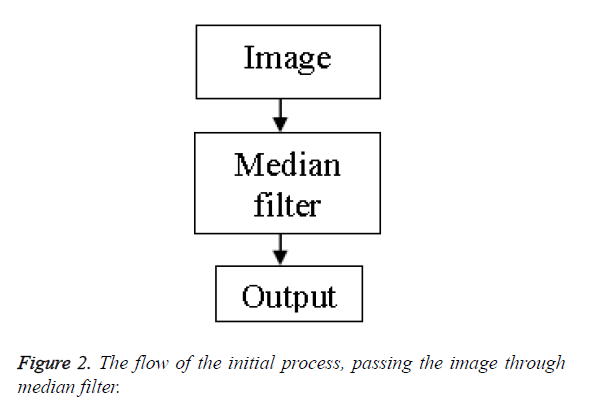
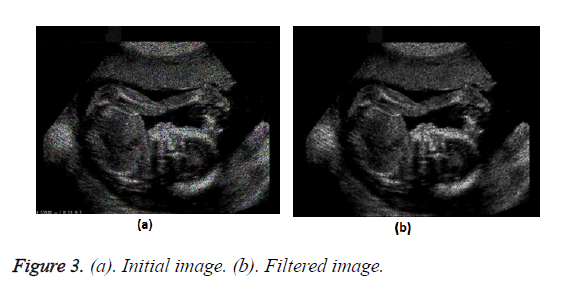
Initial approach: In the initial approach we merely pass the raw unprocessed image through the median filter with the aim to get rid of the speckle noise present. On doing this the output image contains a reduced percentage of speckles in it. However the clarity of the image is not satisfactory.
Figure 2 shows the flow chart of the process. Figure 3 shows the transform from the initial image to the filtered image.
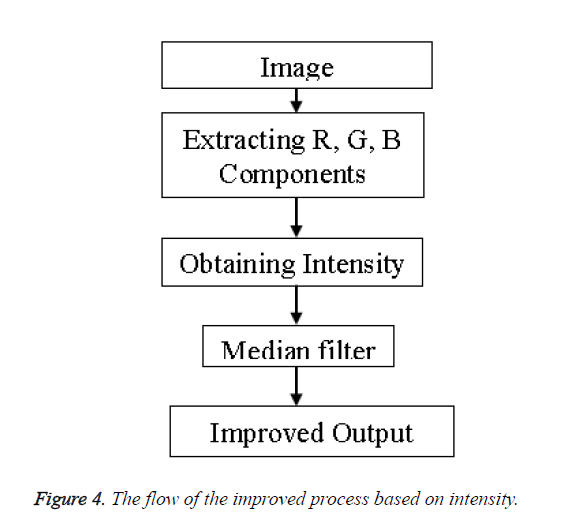
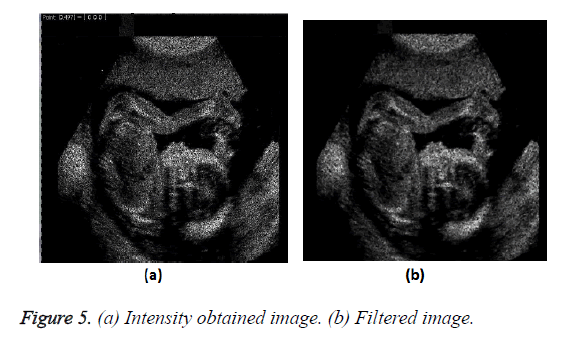
Improved method: The output of the previous method was not very impressive hence to yield better result we incorporate an additional parameter of intensity into the process whereby we first extract each color component of the image i.e. the Red, Green and Blue components (RGB). On obtaining the RGB components the intensity of the image is calculated using the formula of Equation 3.
Intensity=0.3 × Red+0.5 × Green+0.2 × Blue → (3)
The process adopted is shown in the flow chart of Figure 4.
The improved transformation of the image on obtaining the intensity value and passing through the filter is shown in Figure 5.
Proposed method
With the addition of the Intensity parameter the result obtained was satisfactory however to yet reduces the noise of the image in order to yield a better and efficient result. We have proposed a novel method which filters the image with the intensity parameter included in additional parameters and perform segmentation of the image based on the fuzzy c-means concept before passing the image through the filter. The segmentation of the image is performed by the method of clustering. The basic idea in clustering is grouping of objects such that object of one group are similar to one another rather than object of other groups. Clustering can be categorized into: exclusive clustering, hierarchical clustering and overlapping clustering. Fuzzy c-means is an overlapping clustering algorithm.
In the proposed method we initially obtain the intensity scale of the image thereafter grouping every pixel of the image on the concept of fuzzy c-means algorithm. On performing the clusters, each of these clusters are passed through the filter. The filtered result of each cluster is then put together to result in the overall image.
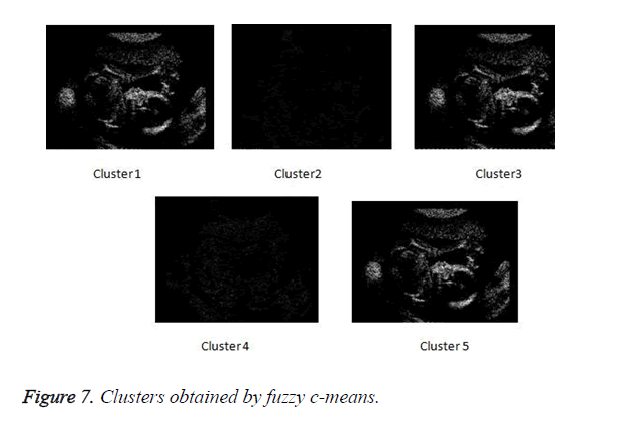
Figure 6 depicts the flow of the process of the proposed method. The clusters obtained by fuzzy c-means clustering.
In Figure 7 the clustered obtained by fuzzy c-means clustering are shown.
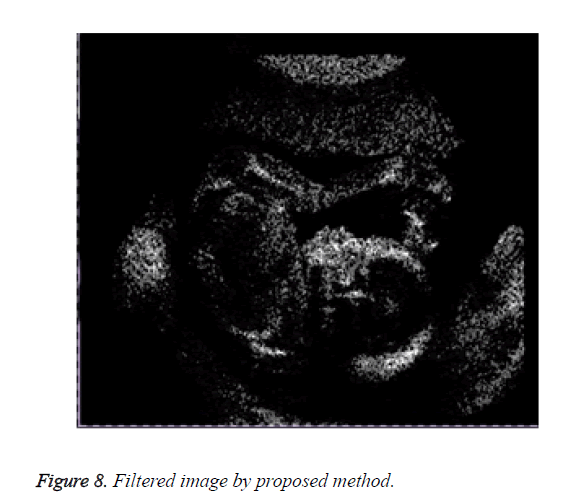
Figure 8 shows the result of the image obtained on applying the proposed method.
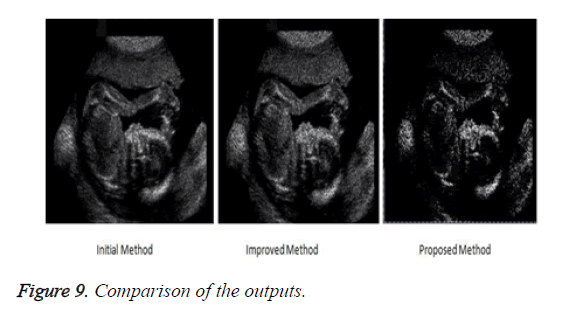
Observation shows that results obtained through our proposed method are better as compared to the initial and improved approaches. Visual inspection being one of the most reliable, strong and efficient testing tool. To support our results we have calculated and compared the outputs of each method against root mean square error measure.
The proposed method produces an image with highly reduced amount of noise as compared to the previous methods. This image when used for medical analysis will produce accurate results and help the diagnosticians to carry out an efficient diagnosis of the patient. Figure 9 compares the result of the filtered image of the three implemented approaches.
The Table 1 below shows the values of three measures used to test the approaches.
| Measures | Initial method filtered image | Improved method filtered image | Proposed method |
|---|---|---|---|
| Mean | 25.1197 | 7.2711 | 6.1224 |
| Standard deviation | 35.3313 | 10.4653 | 8.3145 |
| RMSE | 38.8082 | 31.5343 | 26.1112 |
Table 1. Values obtained for the measures.
Conclusion
On comparing the results it is seen that the proposed method put forth based the concept of clustering and filtering each clustered image yields better results in terms of speckle noise reduction and edge sharpness. For efficient analysis of ultrasound imaging pre-processing is mandatory to reduce the artifacts such as speckle noise present in it. With the reduction of speckle noise the image obtained allows the doctors to perform detail diagnosis which can help in discovering critical information pertaining to health and life.
Future Scope
This study for ultrasound images can be extended to be used with three-Dimensional (3D) ultrasound images. 3D images have an extra element of depth, giving the viewer a virtual reality.
References
- Aaron F, Onal BD, Neale CH. Three-dimensional ultrasound imaging. Phys Med Biol2001; 46: R67-R99.
- Lau LH, Tai F, Yu HL. 3D ultrasound imaging. Univ Hong Kong HKSAR 2010.
- Zhu C, Ni J, Li Y, GuG. Speckle noise suppression techniques for ultrasound images. IEEE InterCompSciEngIntConf 2009.
- Milindkumar VS, Prashant RD. Reduction of speckle noise and image enhancement of images using filtering technique. Int J AdvTechnol 2011; 2:30-38.
- SivakumarR, NedumaranD. Comparative study of speckle noise reduction of ultrasound b-scan images in matrix laboratory environment. Int J CompAppl 2010; 10: 46-50.
- Santosh DHH, Kumar VNL, Ramesh DR, Prasad PS. Efficient techniques for de-noising of speckle noise and highly corrupted impulse noise images. IEEE Electr Comp Technol 2011.